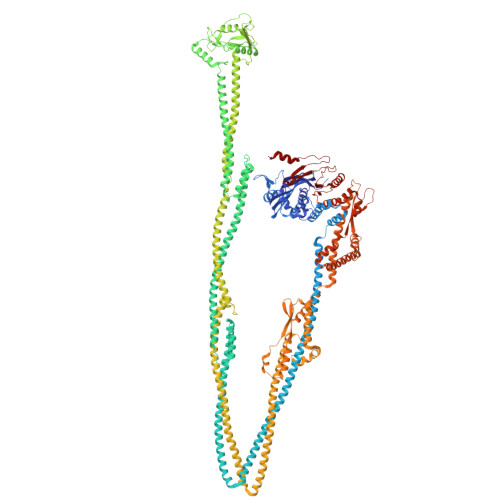
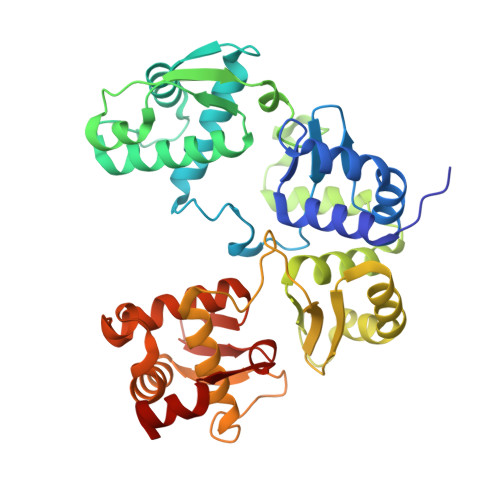
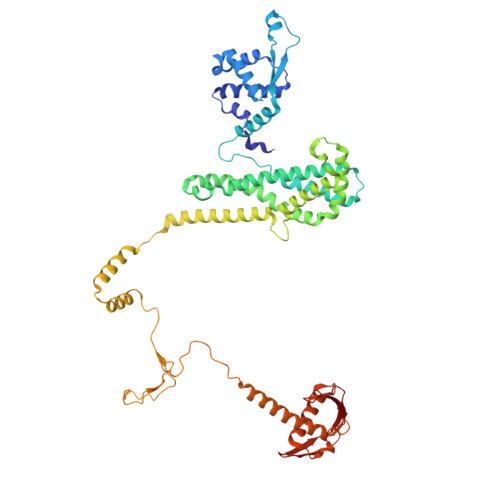
Structure of a type II SMC Wadjet complex from Neobacillus vireti.
Roisne-Hamelin, F., Liu, H.W., Gruber, S.(2025) Structure 33: 1498
- PubMed: 40628265
- DOI: https://doi.org/10.1016/j.str.2025.06.004
- Primary Citation of Related Structures:
9QE0, 9QE1 - PubMed Abstract:
Structural maintenance of chromosome complexes are essential DNA-folding motors that facilitate critical cellular functions, including chromosome segregation and DNA repair. Wadjet systems are prokaryotic SMC complexes specialized in cellular immunity against plasmids. Type I Wadjet systems restrict plasmids via a DNA extrusion-cleavage reaction. Two other Wadjet types (II and III) have also been identified, however, their molecular characteristics are unclear. Here, we reconstituted a representative type II Wadjet system from Neobacillus vireti. We show that this system shares substrate selection and cleavage properties with type I but exhibits distinctive structural features, including a long elbow-distal coiled coil, a channel-less hinge, and a tandem KITE subunit. These features help identify the common and distinguishing architectural elements in the family of Wadjet systems and raise intriguing questions about the evolution of prokaryotic SMC complexes.
- Department of Fundamental Microbiology (DMF), Faculty of Biology and Medicine (FBM), University of Lausanne (UNIL), 1015 Lausanne, Switzerland.
Organizational Affiliation: