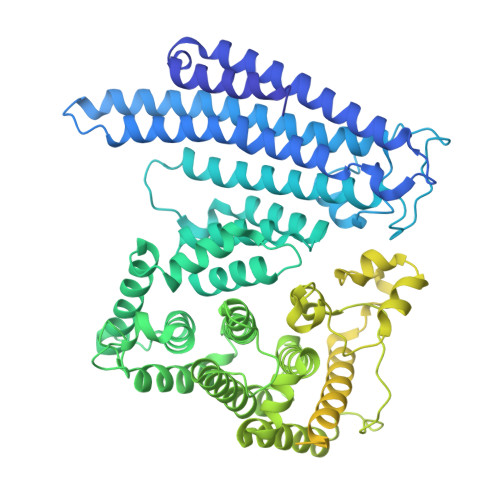
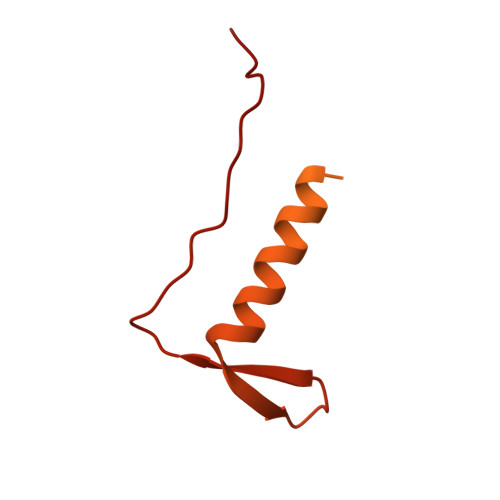
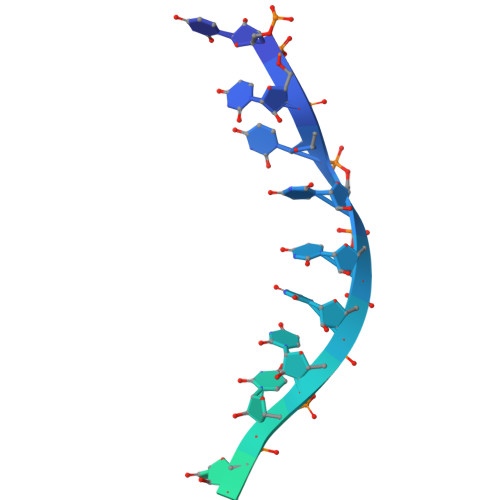
Molecular mechanisms governing the formation of distinct Upf1-containing complexes in yeast.
Iermak, I., Wilson Eisele, N.R., Kurscheidt, K., Loukeri, M.J., Basquin, J., Bonneau, F., Langer, L.M., Keidel, A., Conti, E.(2025) Cell Rep 44: 116415-116415
- PubMed: 41076630
- DOI: https://doi.org/10.1016/j.celrep.2025.116415
- Primary Citation of Related Structures:
9QDQ - PubMed Abstract:
Upf1 is a master regulator of nonsense-mediated mRNA decay (NMD), an mRNA surveillance and degradation pathway conserved from yeast to human. In Saccharomyces cerevisiae, Upf1 exists in two distinct complexes with factors that mediate NMD activation or 5'-3' mRNA degradation. We combined endogenous purifications and biochemical reconstitutions of yeast Upf1 complexes with structural analyses and biochemical assays to elucidate the molecular mechanisms driving the organization of the Upf1-5'-3' and Upf1-2-3 complexes. We show that yeast Upf1 is in a constitutive complex, whereby its CH, RecA, and C-terminal domains interact with the mRNA decapping factor Dcp2, NMD-associated proteins Nmd4 and Ebs1, and the 5'-3' exoribonuclease Xrn1, respectively. Together, the interacting surfaces and closed conformation of Upf1 in the Upf1-5'-3' complex sterically obstruct the binding of Upf2-3. Our work points to a major restructuring upon recruitment of these factors during NMD and provides insights into evolutionary divergence amongst species.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Am Klopferspitz 18, Martinsried, 82152 Munich, Germany. Electronic address: iermak@biochem.mpg.de.
Organizational Affiliation: