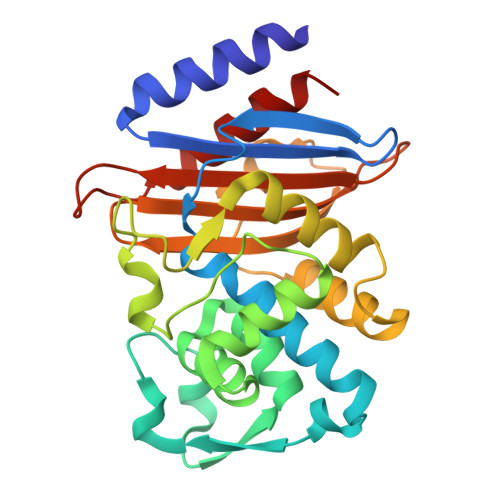
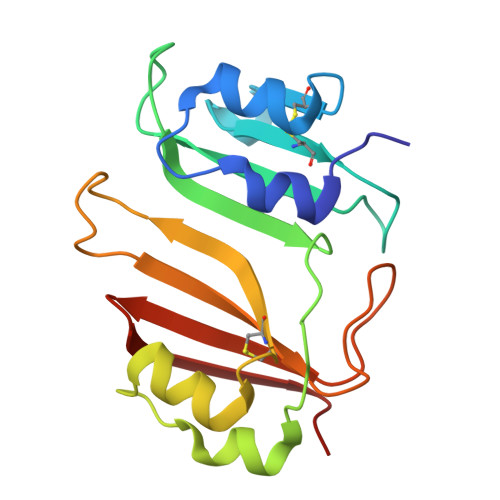
A beta-lactamase inhibitory protein mutant displays high potency and a broad inhibition profile due to an altered binding mode with beta-lactamases.
Rivera, P., Lu, S., Ngango, D., Sankaran, B., Venkataram Prasad, B., Palzkill, T.(2025) J Biological Chem 301: 110850-110850
- PubMed: 41135675
- DOI: https://doi.org/10.1016/j.jbc.2025.110850
- Primary Citation of Related Structures:
9Q0A, 9Q0B, 9Q0C - PubMed Abstract:
β-lactamase enzymes inactivate β-lactam antibiotics, leading to drug resistance. The β-lactamase inhibitory protein (BLIP) is a naturally occurring inhibitor of β-lactamases, with inhibition constants (K i ) ranging from picomolar to micromolar values. For example, BLIP inhibits CTX-M-14 β-lactamase with a K i of 330 nM while the K i for CTX-M-15 is 3 nM, despite CTX-M-14 and CTX-M-15 sharing 83% sequence identity. We used a genetic screen to identify a BLIP mutant, E73W, that potently inhibited CTX-M-14. Subsequent purification and testing of BLIP E73W revealed it is a potent, broad-spectrum inhibitor of class A β-lactamases. We determined structures of BLIP E73W in complex with the CTX-M-14, CTX-M-15, and TEM-1 β-lactamases to investigate the basis of the broad-spectrum inhibition. Previous structures of BLIP in complex with several class-A β-lactamases revealed that β-lactamase active site residue Tyr105 is found in an altered rotamer conformation. Also, in the case of the BLIP/CTX-M-15 complex, an altered conformation of the active site 103-106 loop is observed. In contrast, the BLIP E73W/β-lactamase complexes did not show the altered conformations of Tyr105 or the 103-106 loop. Instead, the mutant's mechanism involves BLIP Trp73 trapping Tyr105 against the wall of the active site in a similar conformation as in the apo-enzyme. Interestingly, the E73W mutant binds the apo-enzyme conformation in all of the BLIP E73W/β-lactamase complexes. Binding to the apo-enzyme conformation, which is expected to be highly populated in solution, as well as enhanced hydrophobic interactions of Trp73 with β-lactamases are possible explanations for the high potency and broad-spectrum inhibition.
- Verna and Marrs McLean Department of Biochemistry and Molecular Pharmacology, Baylor College of Medicine, Houston, TX.
Organizational Affiliation: