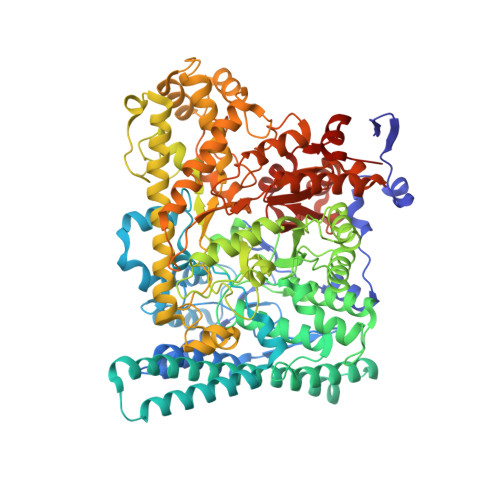
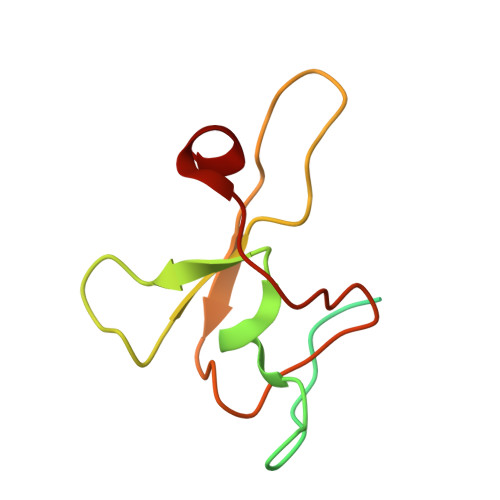
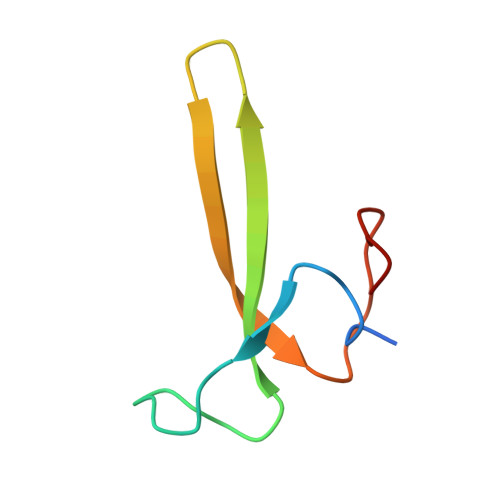
Accessory Subunit Regulates Thiyl Radical Formation in Benzylsuccinate Synthase.
Anas, S., Liu, J., Vats, A., Gainadi, R., Sharif, S., Piriyatamwong, A., Andorfer, M.C.(2025) Biochemistry 64: 4414-4423
- PubMed: 41077954
- DOI: https://doi.org/10.1021/acs.biochem.5c00492
- Primary Citation of Related Structures:
9PZJ - PubMed Abstract:
X-succinate synthases (XSSs) are a class of glycyl radical enzymes (GREs) that enable anaerobic hydrocarbon functionalization, granting anaerobes access to petroleum-derived substrates for metabolism. Owing to their ability to functionalize components of crude oil and catalyze selective olefin hydroalkylation, XSSs hold significant biotechnological promise. However, mechanistic understanding has been limited due to long-standing barriers to installing their essential glycyl radical in vitro, which have only recently been overcome. Unlike most GREs, XSSs contain accessory subunits that bind to the periphery of the catalytic subunit. The most well-studied XSS, benzylsuccinate synthase (BSS), includes two [4Fe-4S] cluster-binding accessory subunits, BSSγ and BSSβ. The full structure of BSSγ and the catalytic role of BSSβ have remained unclear. Here, we report the crystal structure of BSSγ with its [4Fe-4S] cluster intact, revealing a HiPP-like fold similar to that of BSSβ. Through biochemical and spectroscopic studies, we provide evidence that BSSβ promotes thiyl radical formation, even in the absence of a substrate. This finding contrasts with recent models, in which substrate binding is required to trigger thiyl radical formation. With this mechanistic insight, we optimized reaction conditions to achieve total turnover numbers of ∼17,000, representing an over 340-fold improvement compared to prior reports. We further show that in the absence of BSSβ, activated BSSαγ remains catalytically active for up to 11 days. Together, these results clarify the unique regulatory architecture of BSS and lay the groundwork for the use of XSSs in biocatalytic applications.
- Department of Chemistry, Michigan State University, East Lansing, Michigan 48824, United States.
Organizational Affiliation: