Hamster Scap/Insig-2 complex L1-L7 domain/Fab4G10 focused map
Williams, B.C., Kober, D.L., Bai, X., Radhakrishnan, A., Rosenbaum, D.M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sterol regulatory element-binding protein cleavage-activating protein | 797 | Cricetulus griseus | Mutation(s): 7 Gene Names: SCAP | 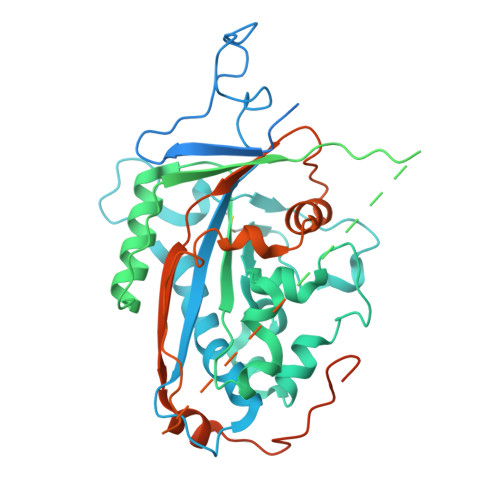 | |
UniProt | |||||
Find proteins for P97260 (Cricetulus griseus) Explore P97260 Go to UniProtKB: P97260 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P97260 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fab4G10 heavy chain | B [auth H] | 252 | Mus musculus | Mutation(s): 0 | 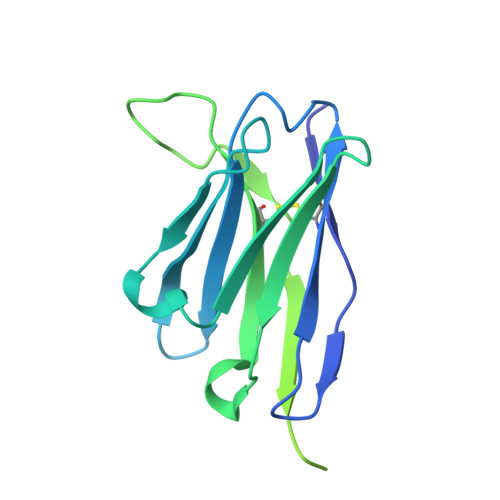 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fab4G10 light chain | C [auth L] | 229 | Mus musculus | Mutation(s): 0 | 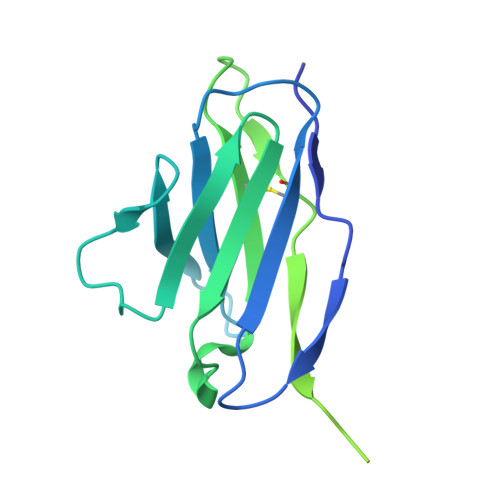 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| RECONSTRUCTION | RELION |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R35GM116387 |
| National Institutes of Health/National Heart, Lung, and Blood Institute (NIH/NHLBI) | United States | P01HL160487 |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | R01AI158357 |