In situ microED structure of the Eosinophil major basic protein-1
Yang, J.E., Bingman, C.A., Mitchell, J., Mosher, D., Wright, E.R.To be published.
Experimental Data Snapshot
Starting Models: experimental, in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bone marrow proteoglycan | A [auth B], B [auth A] | 116 | Homo sapiens | Mutation(s): 0 Gene Names: PRG2, MBP | 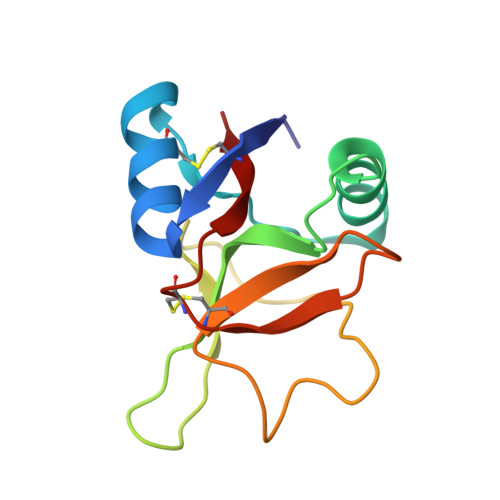 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P13727 (Homo sapiens) Explore P13727 Go to UniProtKB: P13727 | |||||
PHAROS: P13727 GTEx: ENSG00000186652 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13727 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CL (Subject of Investigation/LOI) Query on CL | C [auth B], D [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 30.2 | α = 90 |
| b = 57.8 | β = 91.2 |
| c = 58.6 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PDB_EXTRACT | data extraction |
| PHENIX | phasing |
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.1_5286 |
| RECONSTRUCTION | PHENIX | 1.21.1-5286-000 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01 GM114561 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | U24 GM139168 |
| National Institutes of Health/National Heart, Lung, and Blood Institute (NIH/NHLBI) | United States | P01 HL088594 |
| Department of Energy (DOE, United States) | United States | DE-SC0023013 |
| Department of Energy (DOE, United States) | United States | DE-SC0018409 |