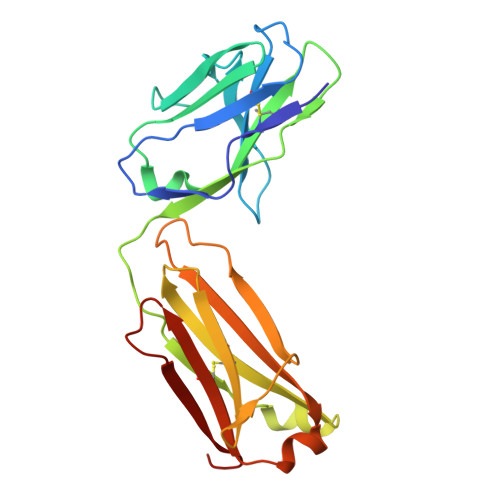
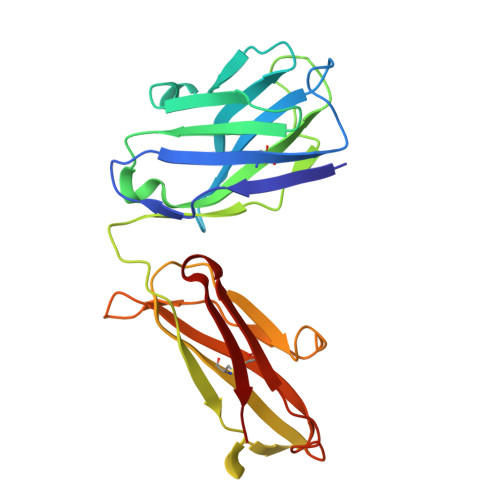
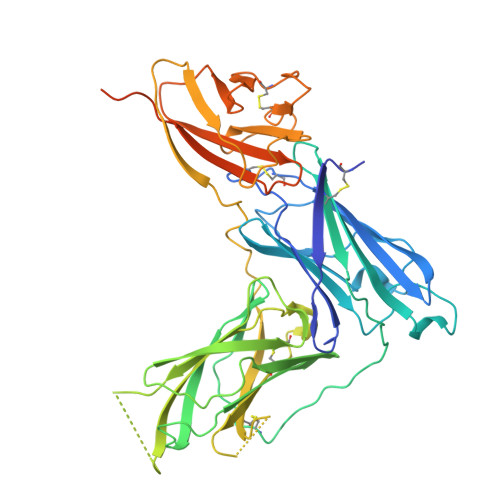
Transgenic Mouse-Derived Human Monoclonal Antibodies Targeting EBV gp350 and gp42 Provide Basis for Therapeutic Development
Chan, C.B., Lang, K., Davis, A.R., Wan, Y.H., Aldridge, N.T., Kher, G., Scharffenberger, S.C., Hardy, S.R., Iureniev, R., Giltiay, N.V., Edwards, K.R., Radtke, S., Kiem, H.P., Pancera, M., McGuire, A.To be published.