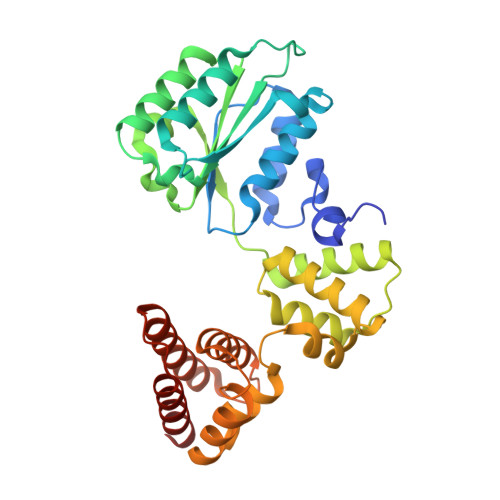
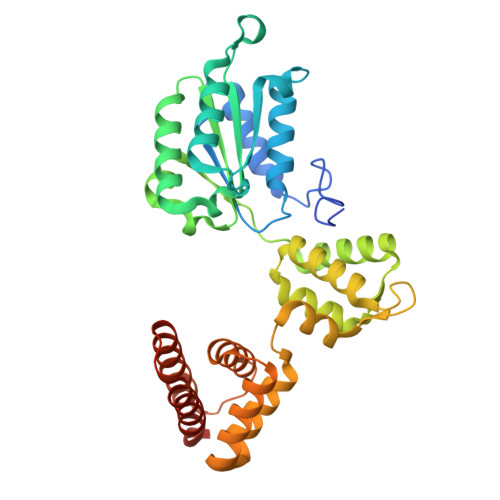
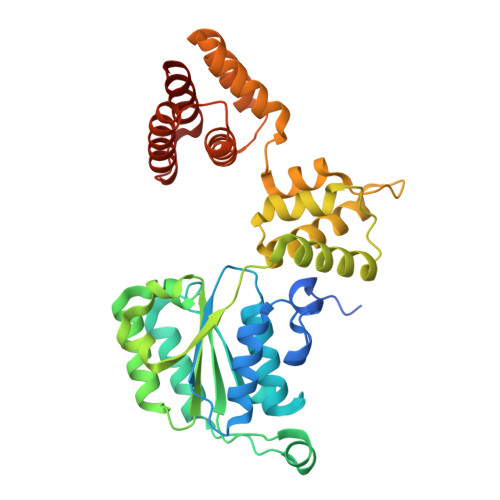
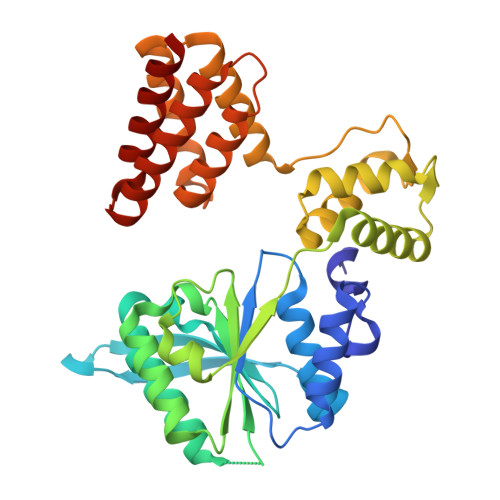
PCNA is a nucleotide exchange factor for the clamp loader ATPase complex.
Pajak, J., Landeck, J.T., Liu, X., Anand, K., Litvak, S., Kelch, B.A.(2025) Proc Natl Acad Sci U S A 122: e2518834122-e2518834122
- PubMed: 41231947
- DOI: https://doi.org/10.1073/pnas.2518834122
- Primary Citation of Related Structures:
9PEO, 9PER, 9PES, 9PET, 9PEU, 9PEV - PubMed Abstract:
All life requires loading ring-shaped sliding clamp protein complexes onto DNA. The sliding clamp loader is a conserved AAA+ ATPase that binds the sliding clamp, opens the ring, and places it onto DNA. While recent structural work on both the canonical and "alternative" clamp loaders has shed light into how these machines perform their task once, it remains unclear how clamp loaders are recycled to load multiple sliding clamps. Here, we present structures of the Saccharomyces cerevisiae clamp loader Replication Factor C (RFC) in absence of sliding clamp or supplemented nucleotide. Our structures indicate that RFC holds onto ADP tightly in at least two of its four ATPase active sites, suggesting that nucleotide exchange is regulated. Our molecular dynamics simulations and biochemical data indicate that binding of the sliding clamp Proliferating Cell Nuclear Antigen (PCNA) causes rapid exchange of tightly bound ADP. Our data suggest that PCNA acts as a nucleotide exchange factor (NEF) by prying apart adjacent subunits, providing a pathway for ADP release. We propose that, by using its own substrate as a NEF, RFC excludes off-pathway states that would arise from binding DNA prior to PCNA.
- Department of Biochemistry and Molecular Biotechnology, University of Massachusetts Chan Medical School, Worcester, MA 01605.
Organizational Affiliation: