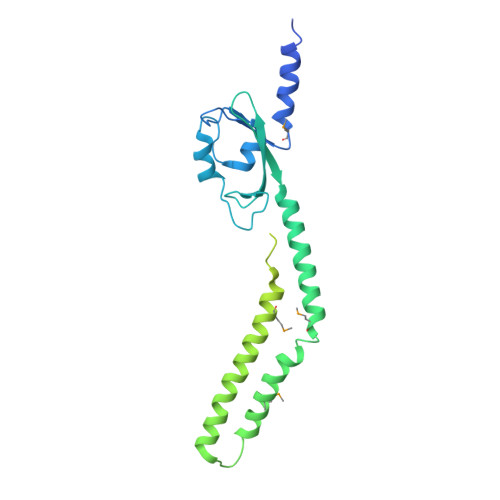
The PAS domain of the polarly localized histidine kinase FlrB in Vibrio cholerae controls class III flagellar transcription and contributes to intestinal colonization.
Stanton, V., Himes, B., Mejia-Santana, A., Eppinger, M., Romo, J., Xing, J., Zhulin, I.B., Cai, H., Wang, Y., Inniss, N.l., Minasov, G., Satchell, K.J.F., Klose, K.E.(2025) mBio 16: e0237925-e0237925
- PubMed: 40981415
- DOI: https://doi.org/10.1128/mbio.02379-25
- Primary Citation of Related Structures:
9P6I - PubMed Abstract:
Vibrio cholerae motility is mediated by a single polar flagellum, composed of four flagellin subunits (FlaABCD) in the filament; however, only FlaA is required for motility. Class III flagellar genes, which include flaA , are controlled by the two-component FlrBC system. FlrB is a histidine kinase that phosphorylates FlrC, which activates Class III promoters. The signal(s) that control phosphotransfer between FlrB and FlrC are unknown. A V. cholerae strain lacking the "non-essential" flagellin genes (Δ flaCEDB ) is non-motile. Selection for spontaneous motile strains resulted in mutations localized to a Per-Arnt-Sim (PAS) domain in the FlrB N-terminus, including the mutation L36F. The X-ray crystal structure of FlrB revealed an asymmetric dimer with a unique fold of the PAS domain. Transcriptome analysis showed that class III transcription is increased with the addition of the L36F PAS mutation to FlrB, while class III gene transcription was eliminated with a mutation at the site of phosphorylation (H135N). H135N prevents phosphorylation of purified FlrB, whereas L36F increases phosphorylation, indicating these mutations represent "off" and "on" forms of FlrB. FlrB localizes to the V. cholerae cell pole, and localization is dependent on the flagellar polar targeting protein FlhF. V. cholerae strains containing either "off" or "on" forms of FlrB were defective for intestinal colonization in infant mice (10- to 40-fold defect). Our results demonstrate that the PAS domain controls FlrB activity and class III flagellar gene expression, and FlrB must switch between inactive and active forms in order for V. cholerae to successfully colonize the intestine. Vibrio cholerae causes the severe diarrheal disease cholera when it colonizes the human intestine. The bacteria are able to swim due to a polar flagellum, and motility is linked to disease, as well as environmental persistence. This study demonstrates that FlrB, a key regulatory protein, localizes to the cell pole and controls flagellar gene transcription via a PAS domain that regulates autophosphorylation. The ability of FlrB to switch between active and inactive forms is critical for motility, as well as intestinal colonization, emphasizing the importance of V. cholerae swimming for its ability to cause disease.
- South Texas Center for Emerging Infectious Diseases and Department of Molecular Microbiology and Immunology, University of Texas at San Antonio, San Antonio, Texas, USA.
Organizational Affiliation: