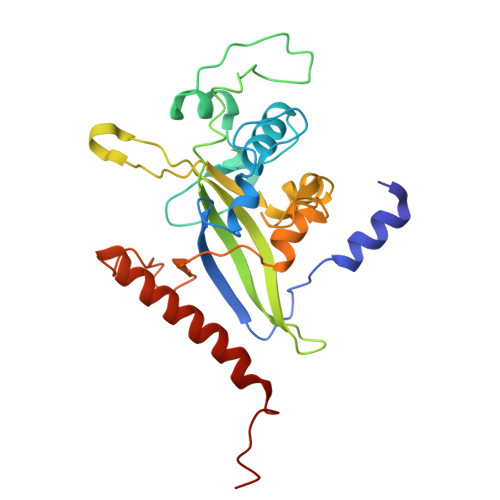
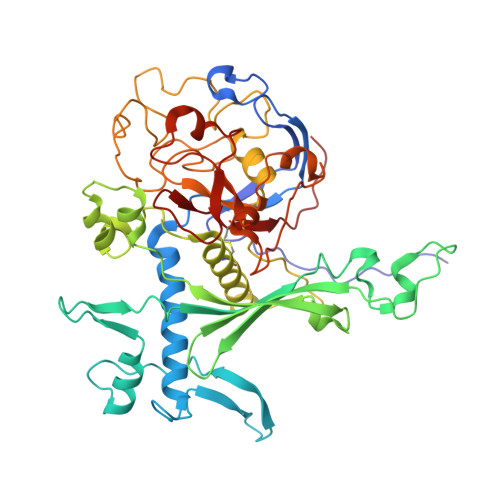
Insights into type IX secretion from PorKN cogwheel structure bound to PorG and attachment complexes.
Gorasia, D.G., Hanssen, E., Mudaliyar, M., Morton, C.J., Valimehr, S., Seers, C., Zhang, L., Doyle, M.T., Ghosal, D., Veith, P.D., Reynolds, E.C.(2025) Nat Commun 16: 7735-7735
- PubMed: 40830366
- DOI: https://doi.org/10.1038/s41467-025-63163-1
- Primary Citation of Related Structures:
9MYJ, 9P6H - PubMed Abstract:
The Type IX Secretion System exports proteins across the outer membrane (OM) of bacteria in the Bacteroidota phylum, however, the mechanistic details remain unknown. In Porphyromonas gingivalis the core components of the multi-protein complex are the Sov translocon, Attachment Complexes (PorQ, U, V, Z), PorLM molecular motors and PorKN rings. Here, we present a ~ 3.5 Å cryo-EM structure of the periplasmic rings comprising 32-33 subunits each of PorK and PorN. Additionally, we show the presence of a critical disulfide bond between PorK and the OM protein PorG that is essential for protein secretion and demonstrate that the Attachment Complexes bind to, and are localized above, the PorKN rings. Overall, each ring resembles a cogwheel with PorN forming cog-like projections that we propose engage with the PorLM motor to drive the rotation of the PorKN cogwheel together with PorG and associated Attachment Complexes, thus providing the energy to complete protein secretion and the coordinated cell surface attachment of the secreted cargo.
- Oral Health Cooperative Research Centre, Melbourne Dental School, The Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Parkville, VIC, Australia. gorasiad@unimelb.edu.au.
Organizational Affiliation: