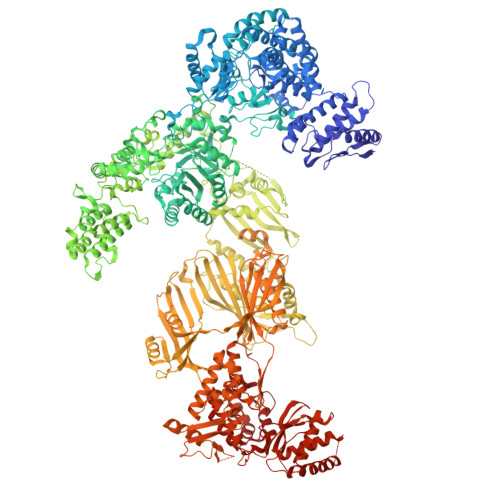
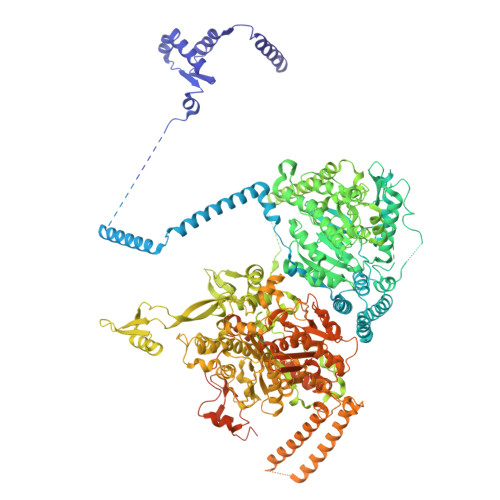
Allosteric regulation of fungal fatty acid synthesis.
Hasan, S.M.N., Samani, E.K., Keszei, A.F.A., Heydari, M., Mazhab-Jafari, M.T.(2025) Structure 33: 2041-2048.e4
- PubMed: 41043416
- DOI: https://doi.org/10.1016/j.str.2025.09.005
- Primary Citation of Related Structures:
9D47, 9D48, 9D49, 9D4A, 9P4V, 9P4W - PubMed Abstract:
Mycobiota fatty acid synthases (FASs) catalyze iterative cycles of condensation, dehydration, and reduction to produce saturated fatty acids. Although these multienzymes are attractive antifungal drug targets, no clinically approved small-molecule inhibitors exist, and the regulation of de novo fatty acid synthesis remains poorly understood. Here, we identify an allosteric regulation of the FAS ketoacyl reduction reaction by palmitoyl-CoA. The palmitate moiety binds a distal site on the central wheel of fungal FAS from Saccharomyces cerevisiae and Candida albicans. This site also accommodates shorter acyl chains, but only palmitoyl-CoA suppresses ketoacyl reductase (KR) activity. While no major conformational changes occur in the reductase domain, palmitoyl-CoA binding quenches dynamics in the central disk, improving local resolution and stabilizing structured water molecules. This entropic effect underlies allosteric communication to the reductase site. Our findings uncover a regulatory mechanism of fungal FAS exploitable for antifungal drug design.
- Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada; Princess Margaret Cancer Center, University Health Network, Toronto, ON, Canada.
Organizational Affiliation: