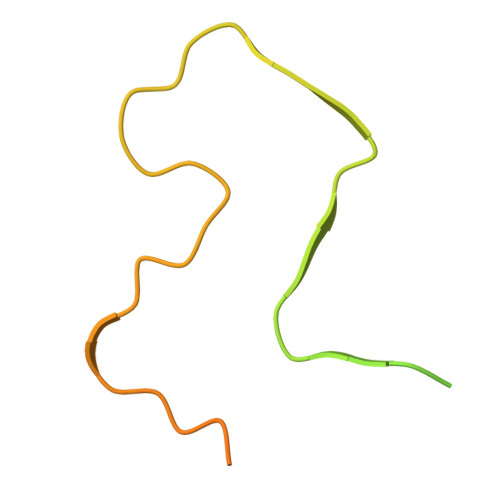
Amyloid fibril structures link CHCHD10 and CHCHD2 to neurodegeneration.
Lv, G., Sayles, N.M., Huang, Y., Mancinelli, C., McAvoy, K., Shneider, N.A., Manfredi, G., Kawamata, H., Eliezer, D.(2025) Nat Commun 16: 7121-7121
- PubMed: 40753073
- DOI: https://doi.org/10.1038/s41467-025-62149-3
- Primary Citation of Related Structures:
9CWW, 9OYO, 9OYQ, 9OYR, 9OYS, 9OYT, 9OYW - PubMed Abstract:
Mitochondrial proteins CHCHD10 and CHCHD2 are mutated in rare cases of heritable FTD, ALS and PD and aggregate in tissues affected by these diseases. Here, we show that both proteins form amyloid fibrils and report cryo-EM structures of fibrils formed from their disordered N-terminal domains. The ordered cores of these fibrils are comprised of a region highly conserved between the two proteins, and a subset of the CHCHD10 and CHCHD2 fibril structures share structural similarities and appear compatible with sequence variations in this region. In contrast, disease-associated mutations p.S59L in CHCHD10 and p.T61I in CHCHD2, situated within the ordered cores of these fibrils, cannot be accommodated by the wildtype structures and promote different protofilament folds and fibril structures. These results link CHCHD10 and CHCHD2 amyloid fibrils to neurodegeneration and further suggest that fibril formation by the WT proteins could also be involved in disease etiology.
- Department of Biochemistry, Weill Cornell Medicine, 1300 York Avenue, New York, NY, USA.
Organizational Affiliation: