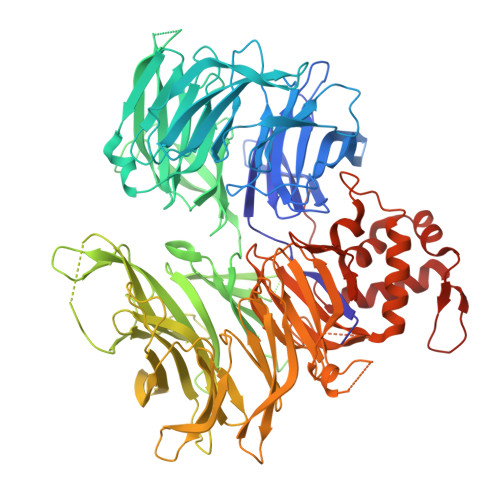
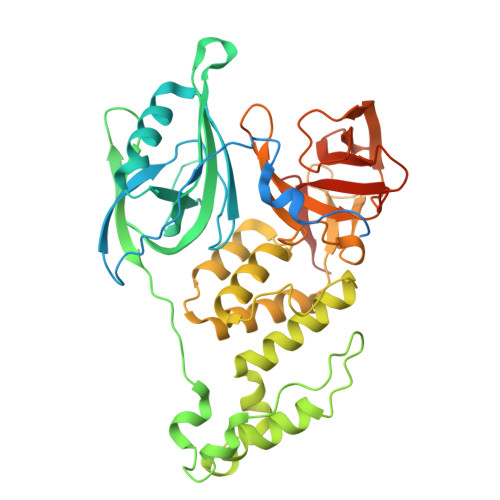
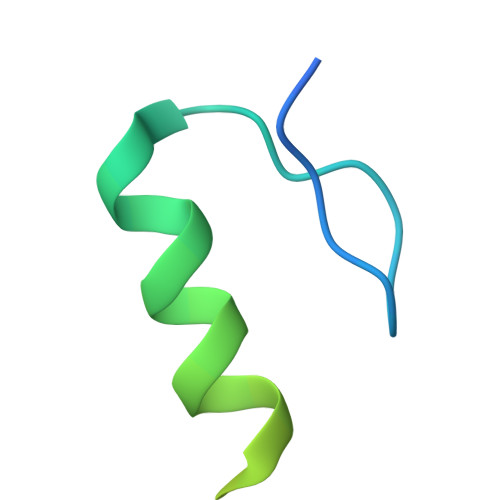
Identification of an allosteric site on the E3 ligase adapter cereblon.
Dippon, V.N., Rizvi, Z., Choudhry, A.E., Chung, C.W., Alkuraya, I.F., Xu, W., Tao, X.B., Jurewicz, A.J., Schneck, J.L., Chen, W., Curnutt, N.M., Kabir, F., Chan, K.H., Queisser, M.A., Musetti, C., Dai, H., Lander, G.C., Benowitz, A.B., Woo, C.M.(2026) Nature
- PubMed: 41565821
- DOI: https://doi.org/10.1038/s41586-025-09994-w
- Primary Citation of Related Structures:
9OTY, 9OUK, 9OUL, 9SFM - PubMed Abstract:
Cereblon (CRBN) is the target of thalidomide derivatives 1 that achieve therapeutic efficacy against some haematologic neoplasias 2-4 by recruiting neosubstrates for degradation 5-7 . Despite the intense investigation of orthosteric thalidomide derivatives, little is known about alternate binding sites on CRBN. Here we report an evolutionarily conserved cryptic allosteric binding site on CRBN. Small-molecule SB-405483 binds the allosteric site to cooperatively enhance the binding of orthosteric ligands and alter their neosubstrate degradation profiles. A survey of over 100 orthosteric ligands and their degradation targets reveals trends in the classes of compounds and neosubstrates in which degradation outcomes are enhanced or inhibited by SB-405483. Structural investigations provide a mechanistic basis for the effects of the allosteric ligand by shifting the conformational distribution of CRBN open to a novel CRBN int and increasing the CRBN closed state. The discovery of a cryptic allosteric binding site on CRBN that alters the functional effects of orthosteric ligands opens new directions with broad implications for improving the selectivity and efficacy of CRBN therapeutics.
- Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, USA.
Organizational Affiliation: