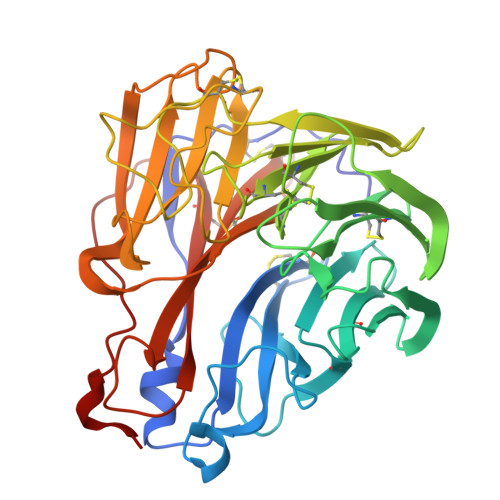
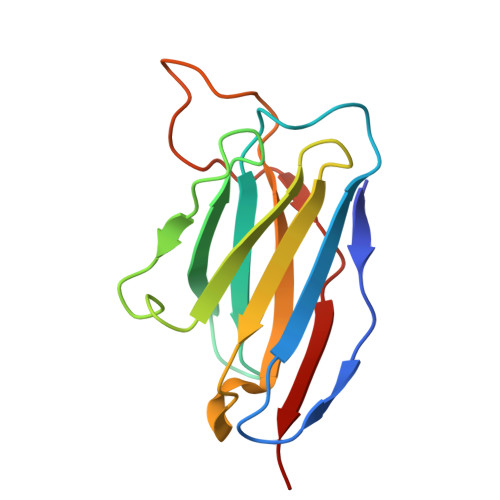
Evolution of antibody cross-reactivity to influenza H5N1 neuraminidase from an N2-specific germline.
Lv, H., Huan, Y.W., Teo, Q.W., Chen, C., Pholcharee, T., Gopal, A.B., Ardagh, M.R., Huang, J.J., Lei, R., Chen, X., Sun, Y., Tang, Y.S., Mehta, A., Szlembarski, M., Mao, K.J., Ma, E.X., Wittenborn, L.E., Tong, M., Rodriguez, L.A., Wang, L., Mok, C.K.P., Wu, N.C.(2025) Cell Host Microbe 33: 1916
- PubMed: 41109220
- DOI: https://doi.org/10.1016/j.chom.2025.09.015
- Primary Citation of Related Structures:
9OSR - PubMed Abstract:
The ongoing spread of highly pathogenic avian influenza H5N1 clade 2.3.4.4b virus in animals and its occasional spillover to humans have raised concerns about a potential H5N1 pandemic. Although recent studies have shown that pre-existing human antibodies can recognize H5N1 neuraminidase, the molecular basis of how this cross-reactivity develops remains poorly understood. In this study, we used a phage display antibody library derived from 245 healthy donors to isolate an antibody, HB420, that cross-reacts with neuraminidases of human H3N2 and avian H5N1 clade 2.3.4.4b viruses and confers protection in vivo. Cryogenic electron microscopy analysis reveals that HB420 targets the neuraminidase active site by mimicking sialic acid binding through a single Asp residue. Furthermore, the inferred germline of HB420 is N2 specific but acquires cross-reactivity to H5N1 neuraminidase through somatic hypermutation. Overall, our findings provide insights into how neuraminidase antibody evolves breadth, which has important implications for the development of broadly protective influenza vaccines.
- Department of Biochemistry, University of Illinois Urbana-Champaign, Urbana, IL 61801, USA; Carl R. Woese Institute for Genomic Biology, University of Illinois Urbana-Champaign, Urbana, IL 61801, USA.
Organizational Affiliation: