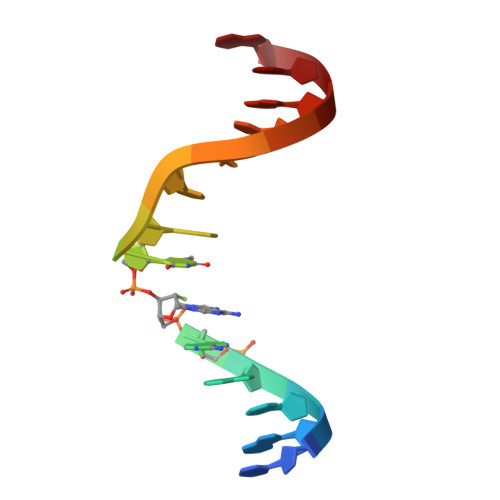
The specificity and structure of DNA cross-linking by the gut bacterial genotoxin colibactin.
Carlson, E.S., Haslecker, R., Lecchi, C., Aguilar Ramos, M.A., Vennelakanti, V., Honaker, L., Stornetta, A., Millan, E.S., Johnson, B.A., Kulik, H.J., Balbo, S., Villalta, P.W., D'Souza, V.M., Balskus, E.P.(2025) Science 390: eady3571-eady3571
- PubMed: 41343624
- DOI: https://doi.org/10.1126/science.ady3571
- Primary Citation of Related Structures:
9OSO - PubMed Abstract:
Accumulating evidence has connected the chemically unstable, DNA-damaging gut bacterial natural product colibactin to colorectal cancer, including the identification of mutational signatures that are thought to arise from colibactin-DNA interstrand cross-links (ICLs). However, we currently lack direct information regarding the structure of this lesion. In this work, we combined mass spectrometry and nuclear magnetic resonance spectroscopy to elucidate the specificity and structure of the colibactin-DNA ICL. We found that colibactin alkylates within the minor groove of adenine- and thymine-rich DNA, explaining the origins of mutational signatures. Unexpectedly, we discovered that the chemically unstable central motif of colibactin mediates the sequence specificity of cross-linking. By directly elucidating colibactin's interactions with DNA, this work enhances our understanding of the structure and genotoxic mechanisms of this cancer-linked gut bacterial natural product.
- Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, USA.
Organizational Affiliation: