Butyrolactol A enhances caspofungin efficacy via flippase inhibition in drug-resistant fungi.
Chen, X., Duan, H.D., Hoy, M.J., Koteva, K., Spitzer, M., Guitor, A.K., Puumala, E., Fiebig, A.A., Hu, G., Yiu, B., Chou, S., Bian, Z., Choi, Y., Guo, A.B.Y., Wang, W., Sun, S., Robbins, N., Averette, A.F., Cook, M.A., Truant, R., MacNeil, L.T., Brown, E.D., Kronstad, J.W., Coombes, B.K., Cowen, L.E., Heitman, J., Li, H., Wright, G.D.(2026) Cell 189: 620
- PubMed: 41478284
- DOI: https://doi.org/10.1016/j.cell.2025.11.036
- Primary Citation of Related Structures:
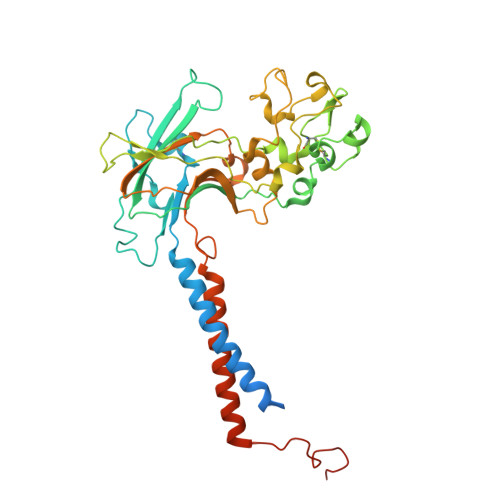
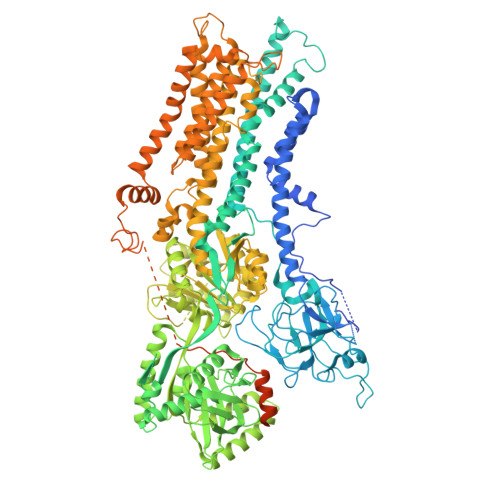
9DZV, 9OMV - PubMed Abstract:
Fungal infections cause millions of deaths annually and are challenging to treat due to limited therapeutic options and rising resistance. Cryptococci are intrinsically resistant to the latest generation of antifungals, echinocandins, while Candida auris, a notorious global threat, is also increasingly resistant. We performed a natural product screen to rescue caspofungin fungicidal activity against Cryptococcus neoformans H99 and identified butyrolactol A, which restores echinocandin efficacy against resistant fungal pathogens, including multidrug-resistant C. auris. Mode-of-action studies reveal that butyrolactol A inhibits the phospholipid flippase Apt1-Cdc50, blocking phospholipid transport. Cryo-electron microscopy analysis of the Apt1-butyrolactol A complex reveals that the flippase is trapped in a dead-end state. Apt1 inhibition disrupts membrane asymmetry, vesicular trafficking, and cytoskeletal organization, thereby enhancing echinocandin uptake and potency. This study identifies lipid flippases as promising antifungal targets and demonstrates the potential of revisiting natural products to expand the antifungal arsenal and combat resistance.
- David Braley Centre for Antibiotics Discovery, M.G. DeGroote Institute for Infectious Disease Research, Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, ON L8S 4K1, Canada.
Organizational Affiliation: