Specificity and mechanism of the double-stranded RNA-specific J2 monoclonal antibody
Bou-Nader, C., Juma, K.M., Bothra, A., Brasington, A.J., Ghirlando, R., Suzuki, M., Garboczi, D.N., Leppla, S.H., Zhang, J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| J2 Fab heavy chain | A [auth C], C [auth A] | 230 | Mus musculus | Mutation(s): 0 | 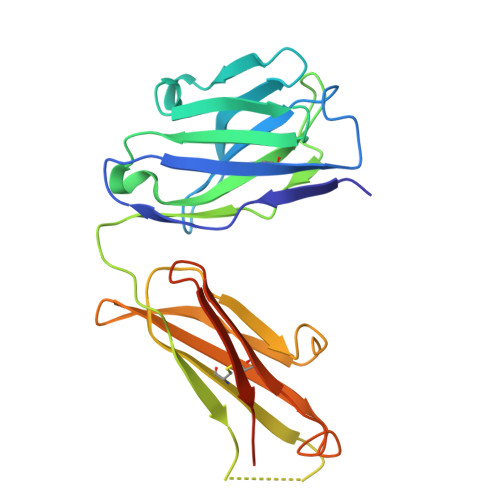 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| J2 Fab light chain | B [auth D], D [auth B] | 219 | Mus musculus | Mutation(s): 0 | 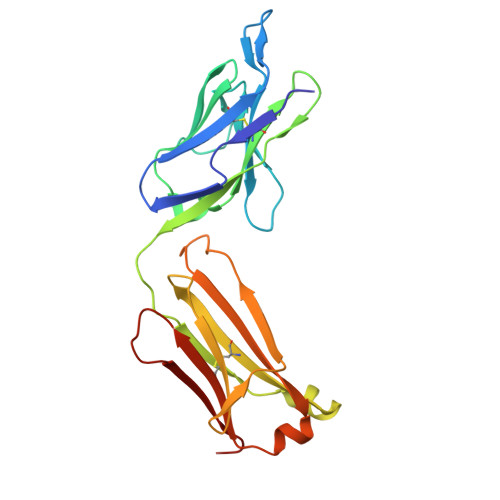 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA strand1 | 24 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA strand2 | 24 | synthetic construct | 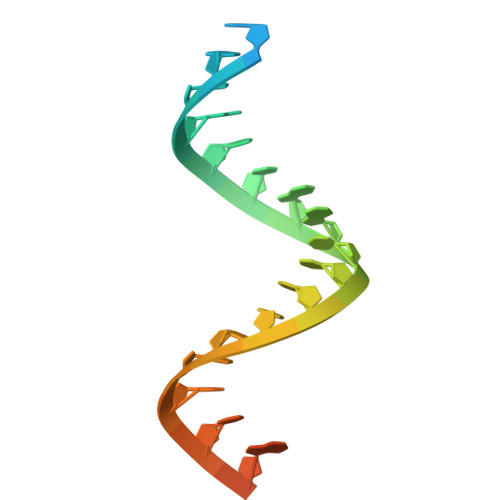 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG (Subject of Investigation/LOI) Query on PEG | G [auth C] H [auth C] I [auth C] J [auth D] K [auth D] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 65.391 | α = 90 |
| b = 92.374 | β = 90 |
| c = 194.663 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Disease (NIH/NIDDK) | United States | -- |