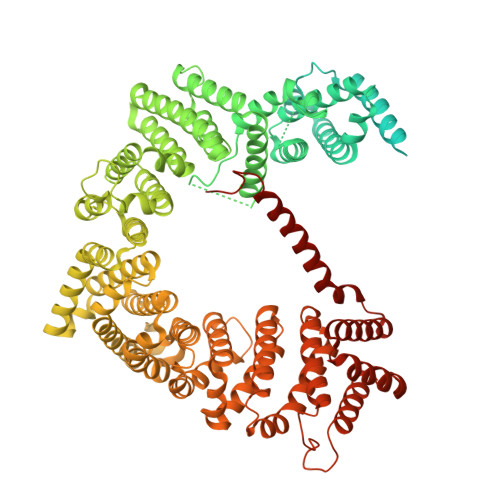
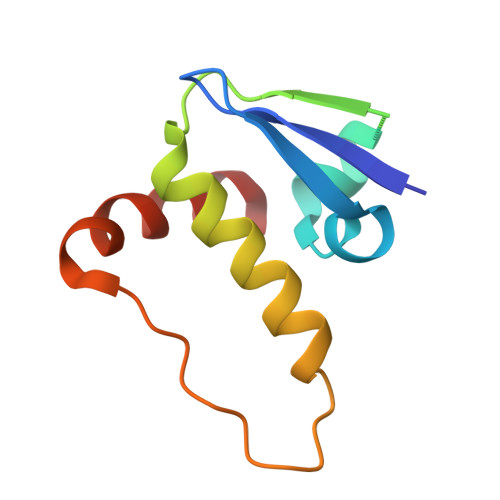
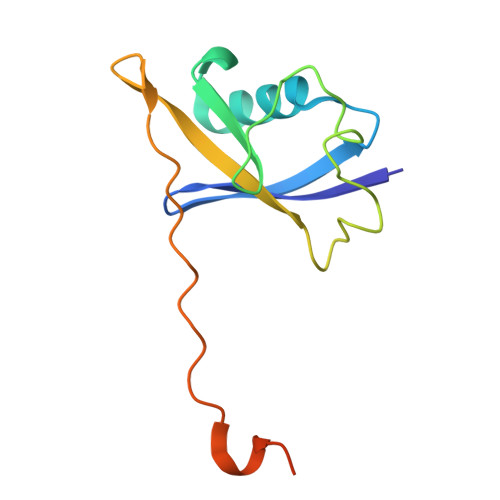
SINE compounds activate exportin-1 degradation via an allosteric mechanism.
Wing, C.E., Fung, H.Y.J., Kwanten, B., Cagatay, T., Niesman, A.B., Jacquemyn, M., Gharghabi, M., Permentier, B., Shakya, B., Nandi, R., Ready, J.M., Kashyap, T., Shacham, S., Landesman, Y., Lapalombella, R., Daelemans, D., Chook, Y.M.(2025) bioRxiv
- PubMed: 39416201
- DOI: https://doi.org/10.1101/2024.10.07.617049
- Primary Citation of Related Structures:
9OG9, 9OGA, 9OGB, 9OGC, 9OGD, 9OGE, 9OGF, 9OGN, 9OGO - PubMed Abstract:
The nuclear export receptor exportin 1 (XPO1/CRM1) is often overexpressed in cancer cells, leading to the mislocalization of numerous cancer-related protein cargoes 1,2 . Selinexor, a covalent XPO1 inhibitor, and other Selective Inhibitor of Nuclear Export (SINEs) restore proper nuclear localization by blocking XPO1-cargo binding 2-7 . SINEs also induce XPO1 degradation via the Cullin-RING E3 ubiquitin ligase (CRL) substrate receptor ASB8 7 . Here we elucidate the mechanism underlying the high-affinity engagement of CRL5 ASB8 with SINE-conjugated XPO1. Cryogenic electron microscopy (cryoEM) structures reveal that ASB8 binds to a cryptic site on XPO1, which becomes accessible only upon SINE conjugation. While molecular glue degraders typically interact with both CRL and the substrate 8-10 , SINEs bind to XPO1 without requiring interaction with ASB8 for efficient XPO1 degradation. Instead, an allosteric mechanism facilitates high affinity XPO1-ASB8 interaction, leading to XPO1 ubiquitination and degradation. ASB8-mediated degradation is also observed upon treatment of the endogenous itaconate derivate 4-octyl itaconate, which suggests a native mechanism that is inadvertently exploited by synthesized XPO1 inhibitors. This allosteric XPO1 degradation mechanism of SINE compounds expands the known modes of targeted protein degradation beyond the well-characterized molecular glue degraders and proteolysis targeting chimeras of CRL4.