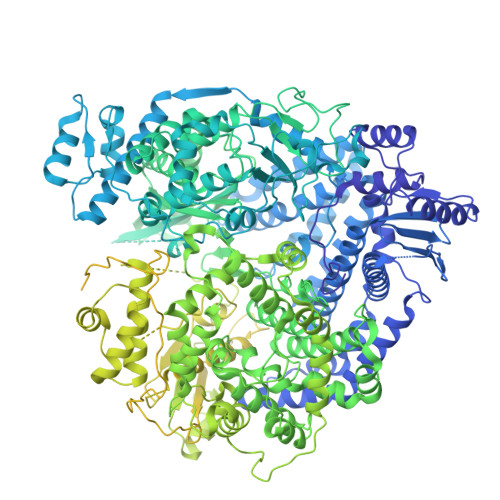
Structural basis of measles virus polymerase inhibition by nonnucleoside inhibitor ERDRP-0519.
Wang, D., Bu, F., Yang, G., Liu, B.(2025) Nat Commun 16: 9061-9061
- PubMed: 41083444
- DOI: https://doi.org/10.1038/s41467-025-64128-0
- Primary Citation of Related Structures:
9OCE, 9OCF - PubMed Abstract:
ERDRP-0519 is a potent nonnucleoside inhibitor active against measles virus (MeV) and other Morbilliviruses. Here we report cryo-EM structures of the compound bound to MeV polymerase complexes at 2.73 Å and 2.48 Å resolution, revealing a unique binding pocket in the RdRp palm subdomain that overlaps the catalytic GDN motif. These findings clarify the basis of resistance mutations, including W671, and provide a foundation for designing next-generation Paramyxovirus antivirals.
- Section of Transcription & Gene Regulation, The Hormel Institute, University of Minnesota, Austin, MN, USA.
Organizational Affiliation: