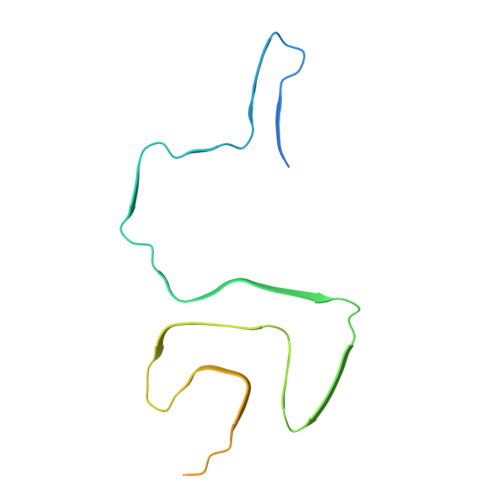
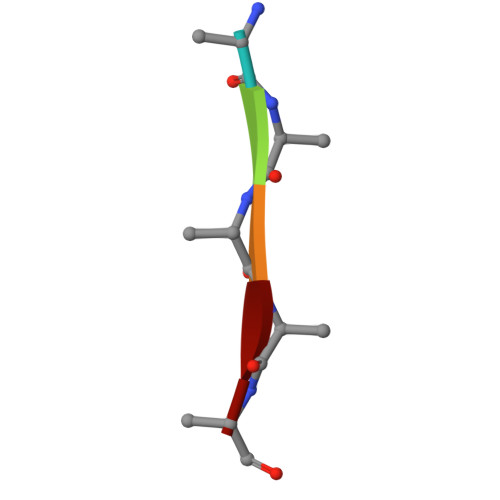
Lewy-MSA hybrid fold drives distinct neuronal alpha-synuclein pathology.
Enomoto, M., Martinez-Valbuena, I., Forrest, S.L., Xu, X., Munhoz, R.P., Li, J., Rogaeva, E., Lang, A.E., Kovacs, G.G.(2025) Commun Biol 8: 929-929
- PubMed: 40523906
- DOI: https://doi.org/10.1038/s42003-025-08355-7
- Primary Citation of Related Structures:
9E9X, 9OBP - PubMed Abstract:
The ordered assembly of α-synuclein protein encoded by SNCA into filaments characterizes neurodegenerative synucleinopathies. Lewy body disease (LBD) shows predominantly neuronal and multiple system atrophy (MSA), predominantly oligodendrocytic α-synuclein pathology affecting subcortical brain structures. Based on cryo-electron microscopy, it was reported that the structures of α-synuclein filaments from LBD differ from MSA and juvenile-onset synucleinopathy (JOS). The rare atypical MSA subtype shows abundant neuronal argyrophilic α-synuclein inclusions in the limbic system. Current concepts indicate that disease entities are characterized by unique protofilament folds. Here we demonstrate that α-synuclein can form a Lewy-MSA hybrid fold, leading to the atypical histopathological form of MSA. Distinct biochemical characteristics of α-synuclein, as demonstrated by protease-sensitivity digestion assay, seed amplification assays (SAAs), and conformational stability assays (CSA), are also linked to cytopathological differences. We expand the current structure-based classification of α-synucleinopathies and propose that cell-specific protein pathologies can be associated with distinct filament folds.
- Princess Margaret Cancer Centre, University Health Network, Toronto, ON, Canada. Masahiro.Enomoto@uhn.ca.
Organizational Affiliation: