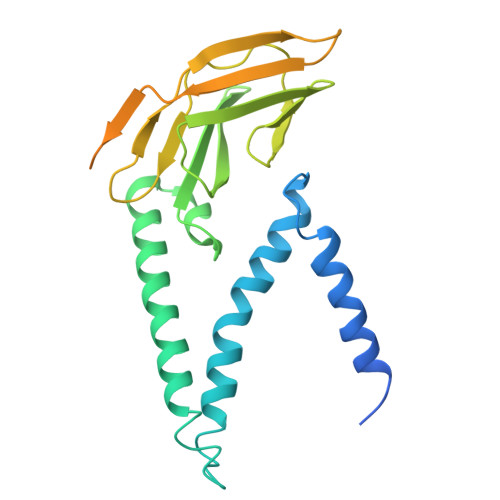
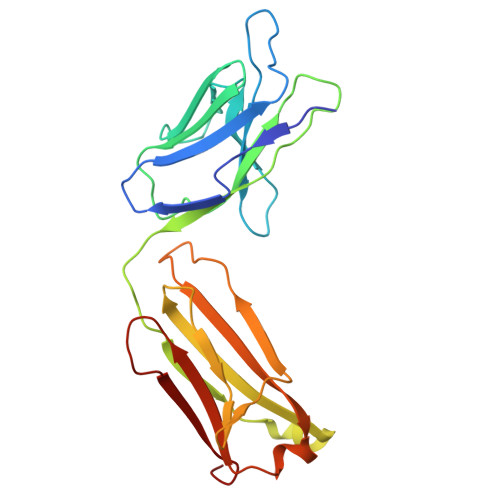
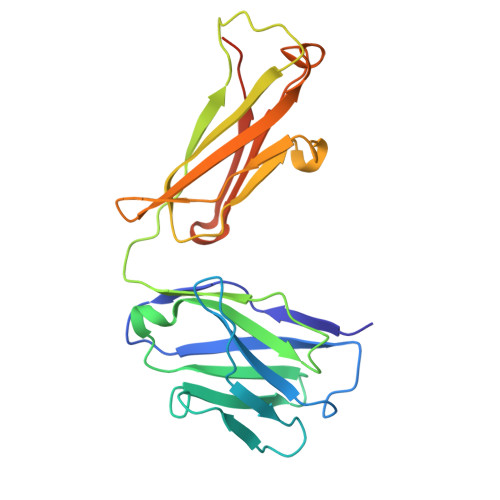
Structural insights into MERS and SARS coronavirus membrane proteins.
Mann, M.K., Yin, Y., Marsili, S., Xie, J., Doijen, J., Miller, R., Piassek, M., van den Broeck, N., Kariuki, C.K., de Gruyter, H.L.M., Leijs, A.A., Snijder, E.J., van Hemert, M.J., Keustermans, K., Van Gool, M., Yu, X., Loock, M.V., Koul, A., Sharma, S., Van Damme, E., Abeywickrema, P.(2025) Commun Biol 8: 1651-1651
- PubMed: 41286109
- DOI: https://doi.org/10.1038/s42003-025-09042-3
- Primary Citation of Related Structures:
9NZ3, 9NZ4, 9NZ5 - PubMed Abstract:
The membrane (M) protein of coronaviruses is essential for maintaining structural integrity during membrane virion budding and viral pathogenesis. Given its high conservation in lineages within the betacoronavirus genus, such as sarbecoviruses, the M protein presents as an attractive therapeutic target; however, developing broad-spectrum antivirals targeting coronaviruses such as MERS-CoV is challenging due to lower sequence conservation and limited structural information available beyond that of the SARS-CoV-2 M protein. In this study, we report 3-3.2 Å resolution structures of MERS-CoV M protein, engineered with a SARS-CoV-2-like antibody interface, representing the first human merbecovirus M protein structure, and SARS-CoV M protein structures, with and without a previously identified SARS-CoV-2 M protein inhibitor, JNJ-9676. We highlight the structural differences between the MERS-CoV, SARS-CoV and SARS-CoV-2 M proteins, and present insights into the conservation of the JNJ-9676 binding pocket as well as key differences that could be targeted to accelerate the design of specific MERS-CoV and broad-spectrum antivirals targeting coronavirus M proteins.
- Johnson and Johnson, Spring House, PA, USA.
Organizational Affiliation: