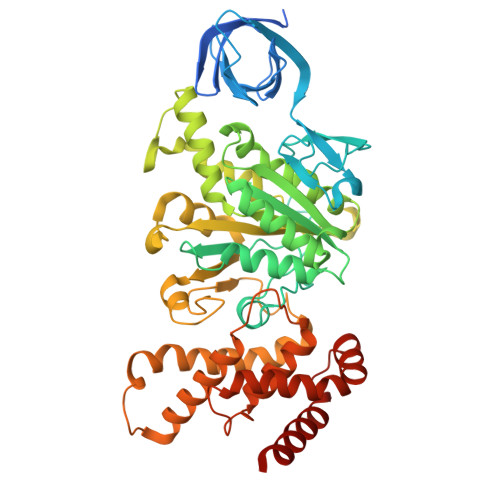
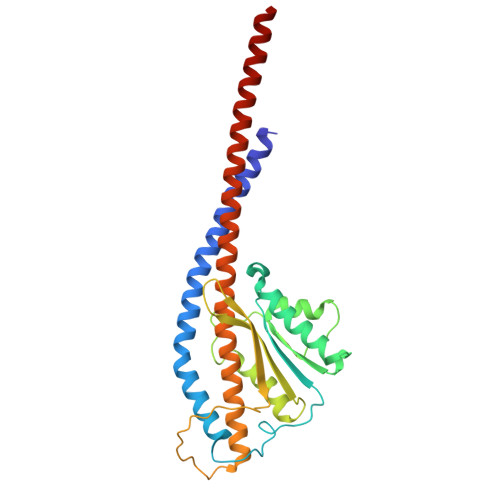
Functional and structural characterization of F 1 -ATPase with common ancestral core domains in stator ring.
Suzuki, A.K., Furukawa, R., Sobti, M., Brown, S.H.J., Stewart, A.G., Akanuma, S., Ueno, H., Noji, H.(2025) Protein Sci 34: e70345-e70345
- PubMed: 41131942
- DOI: https://doi.org/10.1002/pro.70345
- Primary Citation of Related Structures:
9NVL, 9NVM - PubMed Abstract:
Extant F 1 -ATPases exhibit diverse rotational stepping behaviors-3-, 6-, or 9-step cycles-yet the evolutionary origin of these patterns remains unclear. Here, we used ancestral sequence reconstruction to infer the catalytic β and non-catalytic α subunits of a putative ancestral F 1 -ATPase. We then fused their functionally critical domains into the thermostable F 1 from Bacillus PS3, yielding a stable chimeric enzyme. Cryo-EM revealed two distinct conformational states-binding and catalytic dwell states-separated by a ~34° rotation of the γ subunit, suggesting a fundamental six-step mechanism akin to that of extant six-stepping F 1 -ATPases. Single-molecule rotation assays with ATP and the slowly hydrolyzed ATP analog ATPγS demonstrated that the chimeric motor is intrinsically a six-stepper, pausing at binding and catalytic dwell positions separated by 32.1°, although the binding dwell is significantly prolonged by an unknown mechanism. These findings indicate that F 1 -ATPase was originally a six-stepper and diversified into 3-, 6- and 9-step forms in evolutionary adaptation. Based on these results, we discuss plausible features of the entire F o F 1 complex, along with potential physiological contexts in the last universal common ancestor and related lineages.
- Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: