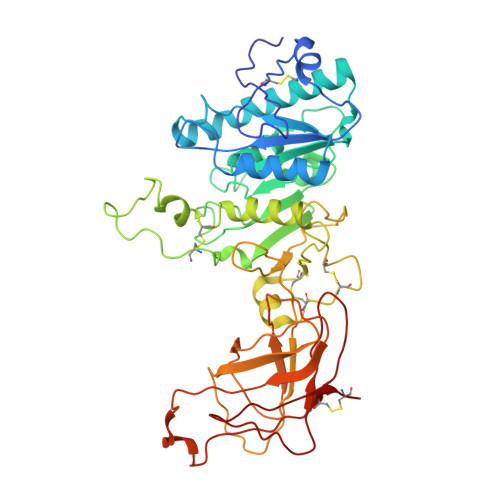
Cryogenic electron tomography reveals helical organization of lipoprotein lipase in storage vesicles.
Gunn, K.H., Wheless, A., Calcraft, T., Kreutzberger, M., El-Houshy, K., Egelman, E.H., Rosenthal, P.B., Neher, S.B.(2025) Sci Adv 11: eadx8711-eadx8711
- PubMed: 40768583
- DOI: https://doi.org/10.1126/sciadv.adx8711
- Primary Citation of Related Structures:
9NRN - PubMed Abstract:
Lipoprotein lipase (LPL) is a triglyceride lipase that is contained in intracellular vesicles in an inactive storage form before secretion, but the precise structural details have not yet been resolved. Using cryo-electron tomography (cryo-ET), we observe that LPL exists inside of storage vesicles as a filament with an 11-nanometer diameter and is packed in these vesicles in two distinct patterns. Next, we solved a 4.2-Å resolution cryo-electron microscopy (cryo-EM) structure of this 11-nanometer LPL filament using purified protein. The filament is made of repeating pairs of LPL molecules with occluded active sites, rendering the LPL inactive. The comparison of the in situ subtomogram average and the in vitro cryo-EM structure indicates that the previously uncharacterized physiological storage form of LPL is an inactive filament.
- Department of Biochemistry and Cell Biology, Stony Brook University, Stony Brook, NY 11790, USA.
Organizational Affiliation: