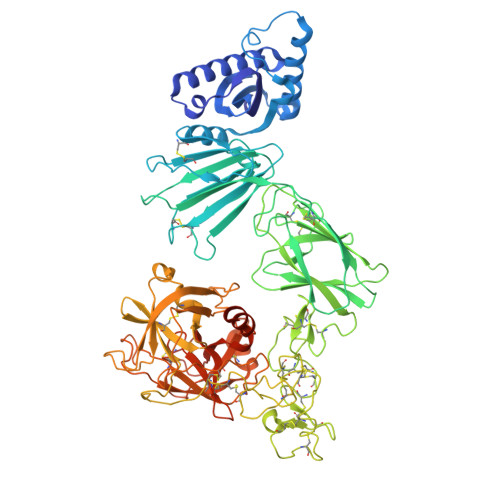
A TMPRSS6-inhibiting mAb improves disease in a beta-thalassemia mouse model and reduces iron in healthy humans.
Lob, H.E., Singh, N., Mohammadi, K., Ivanova, L., Crowell, B., Kim, H.J., Kravets, L., Das, N.M., Ray, Y., Kim, J.H., Rottey, S., Labriola-Tompkins, E., Hassan, H.E., Farrelly, L., Chin, H.F., Preda, M., Spencer Noakes, L., Saotome, K., Franklin, M., Retter, M.W., Karayusuf, E., Flanagan, J.J., Olson, W., Nannuru, K.C., Idone, V., Burczynski, M.E., Harari, O.A., Perlee, L., Van Lancker, G., Murphy, A.J., Economides, A.N., Hatsell, S.J.(2025) JCI Insight 10
- PubMed: 40548380
- DOI: https://doi.org/10.1172/jci.insight.191813
- Primary Citation of Related Structures:
9NRC - PubMed Abstract:
β-Thalassemia is a genetic disorder arising from mutations in the β-globin gene, leading to ineffective erythropoiesis and iron overload. Ineffective erythropoiesis, a hallmark of β-thalassemia, is an important driver of iron overload, which contributes to liver fibrosis, diabetes, and cardiac disease. Iron homeostasis is regulated by the hormone hepcidin; BMP6/hemojuvelin-mediated (BMP6/HJV-mediated) signaling induces hepatic hepcidin expression via SMAD1/5, with transmembrane serine protease 6 (TMPRSS6) being a negative regulator of HJV. Individuals with loss-of-function mutations in the TMPRSS6 gene show increased circulating hepcidin and iron-refractory iron-deficiency anemia, suggesting that blocking TMPRSS6 may be a viable strategy to elevate hepcidin levels in β-thalassemia. We generated a human mAb (REGN7999) that inhibits TMPRSS6. In an Hbbth3/+ mouse model of β-thalassemia, REGN7999 treatment led to significant reductions in liver iron, reduced ineffective erythropoiesis, and showed improvements in RBC health, running distance during forced exercise, and bone density. In a phase I, doubleblind, randomized, placebo-controlled study in healthy human volunteers (NCT05481333), REGN7999 increased serum hepcidin and reduced serum iron with an acceptable tolerability profile. Our results suggest that, by both reducing iron and improving RBC function, inhibition of TMPRSS6 by REGN7999 may offer a therapy for iron overload and impaired erythropoiesis in β-thalassemia.
- Regeneron Pharmaceuticals, Tarrytown, New York, USA.
Organizational Affiliation: