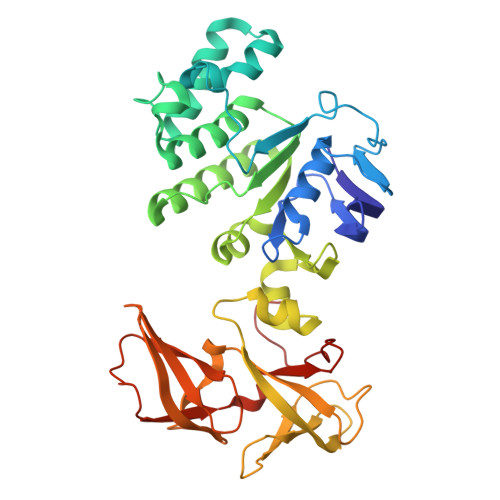
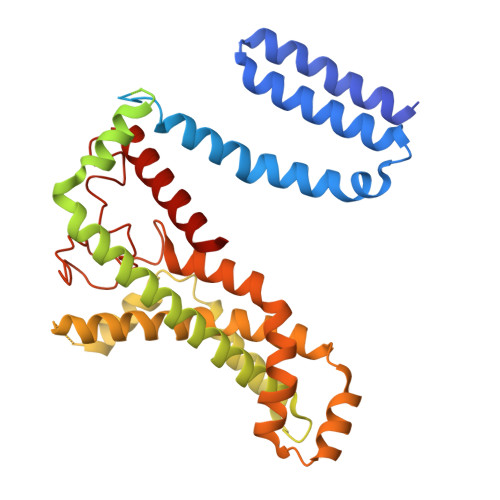
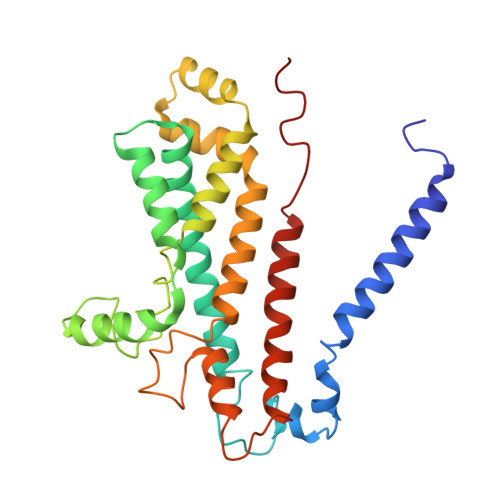
DeFrND: detergent-free reconstitution into native nanodiscs with designer membrane scaffold peptides.
Ren, Q., Wang, J., Idikuda, V., Zhang, S., Shin, J., Ludlam, W.G., Real Hernandez, L.M., Zdancewicz, S., Kreutzberger, A.J.B., Chang, H., Kiessling, V., Tamm, L.K., Jomaa, A., Levental, I., Martemyanov, K., Chanda, B., Bao, H.(2025) Nat Commun 16: 7973-7973
- PubMed: 40858559
- DOI: https://doi.org/10.1038/s41467-025-63275-8
- Primary Citation of Related Structures:
9BCR, 9NQJ, 9NXC - PubMed Abstract:
Membrane scaffold protein-based nanodiscs have facilitated unprecedented structural and biophysical analysis of membrane proteins in a near-native lipid environment. However, successful reconstitution of membrane proteins in nanodiscs requires prior solubilization and purification in detergents, which may impact their physiological structure and function. Furthermore, the detergent-mediated reconstitution of nanodiscs is unlikely to recapitulate the precise composition or asymmetry of native membranes. To circumvent this fundamental limitation of traditional nanodisc technology, we herein describe the development of membrane-solubilizing peptides to directly extract membrane proteins from native cell membranes into nanoscale discoids. By systematically protein engineering and screening, we create a class of chemically modified Apolipoprotein-A1 mimetic peptides to enable the formation of detergent-free nanodiscs with high efficiency. Nanodiscs generated with these engineered membrane scaffold peptides are suitable for obtaining high-resolution structures using single-particle cryo-EM with native lipids. To further highlight the versatility of our approach, we directly extract a sampling of membrane signaling proteins with their surrounding native membranes for biochemical and biophysical interrogations.
- Department of Molecular Medicine, UF Scripps Biomedical Research, Jupiter, Florida, USA.
Organizational Affiliation: