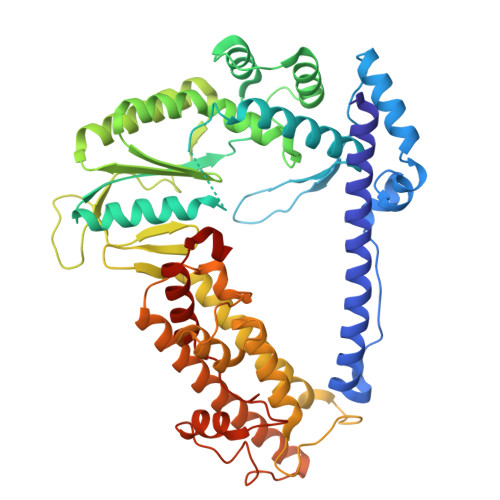
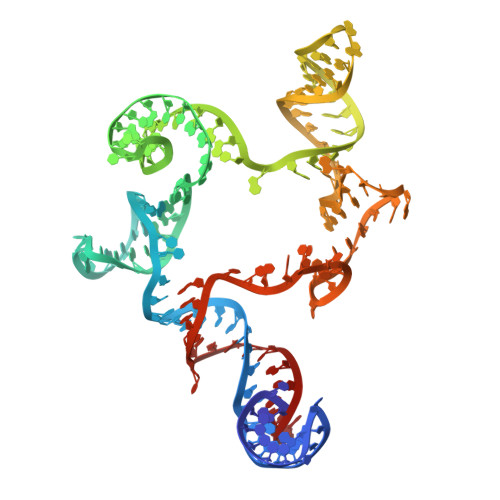
Protein-primed homopolymer synthesis by an antiviral reverse transcriptase.
Tang, S., Zedaveinyte, R., Burman, N., Pandey, S., Ramirez, J.L., Kulber, L.M., Wiegand, T., Wilkinson, R.A., Ma, Y., Zhang, D.J., Lampe, G.D., Berisa, M., Jovanovic, M., Wiedenheft, B., Sternberg, S.H.(2025) Nature 643: 1352-1362
- PubMed: 40436039
- DOI: https://doi.org/10.1038/s41586-025-09179-5
- Primary Citation of Related Structures:
9NLV, 9NLX - PubMed Abstract:
Bacteria defend themselves from viral predation using diverse immune systems, many of which target foreign DNA for degradation 1 . Defense-associated reverse transcriptase (DRT) systems provide an intriguing counterpoint to this strategy by leveraging DNA synthesis instead 2,3 . We and others recently showed that DRT2 systems use an RNA template to assemble a de novo gene that encodes an antiviral effector protein, Neo 4,5 . It remains unknown whether similar mechanisms of defense are employed by other related DRT families. Focusing on DRT9, here we uncover an unprecedented mechanism of DNA homopolymer synthesis. Viral infection triggers polydeoxyadenylate (poly-dA) accumulation in the cell, driving abortive infection and population-level immunity. Cryo-EM structures reveal how a noncoding RNA serves as both a structural scaffold and reverse transcription template to direct hexameric complex assembly and poly-dA synthesis. Remarkably, biochemical and functional experiments identify tyrosine residues within the reverse transcriptase itself that likely prime DNA synthesis, leading to the formation of high-molecular weight protein-DNA covalent adducts. Synthesis of poly-dA by DRT9 in vivo is regulated by the competing activities of phage-encoded triggers and host-encoded silencers. Collectively, our work unveils a novel nucleic acid-driven defense system that expands the paradigm of bacterial immunity and broadens the known functions of reverse transcriptases.
- Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA.
Organizational Affiliation: