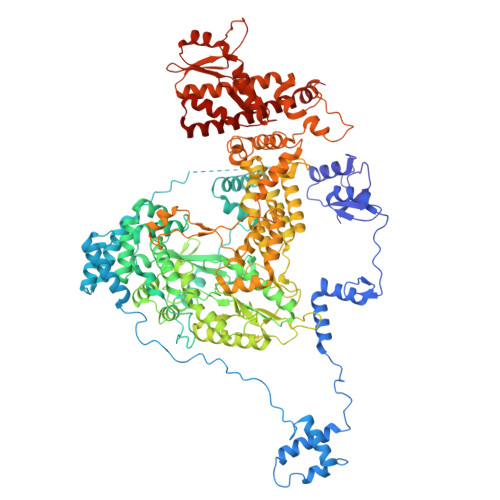
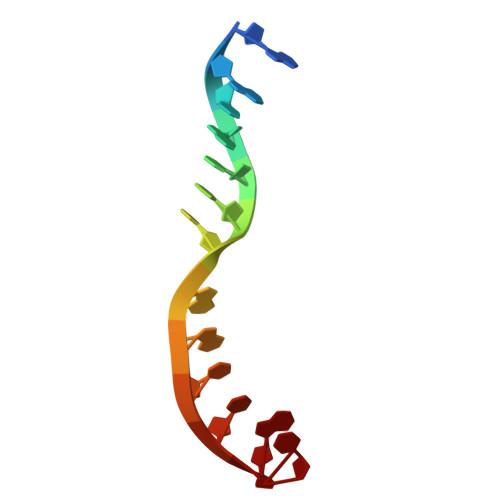
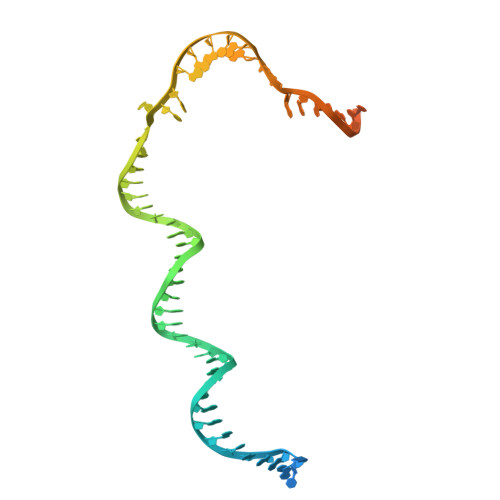
Structures of vertebrate R2 retrotransposon complexes during target-primed reverse transcription and after second-strand nicking.
Thawani, A., Rodriguez-Vargas, A., Van Treeck, B., Hassan, N.T., Adelson, D.L., Nogales, E., Collins, K.(2025) Sci Adv 11: eadu5533-eadu5533
- PubMed: 40540573
- DOI: https://doi.org/10.1126/sciadv.adu5533
- Primary Citation of Related Structures:
9NL2, 9NL3, 9NL4 - PubMed Abstract:
R2 retrotransposons are site-specific eukaryotic non-long terminal repeat retrotransposons that copy and paste into gene loci encoding ribosomal RNAs. Recently, we demonstrated that avian A-clade R2 proteins achieve efficient and precise insertion of transgenes into their native safe-harbor loci in human cells. The features of A-clade R2 proteins that support gene insertion are not well characterized. Here, we report high-resolution cryo-electron microscopy structures of two vertebrate A-clade R2 proteins at the initiation of target-primed reverse transcription and after cDNA synthesis and second-strand nicking. Using biochemical and cellular assays, we illuminate the basis for high selectivity of template use and unique roles for each of the three zinc-finger domains in nucleic acid recognition. Reverse transcriptase active site architecture is reinforced by an unanticipated insertion motif specific to vertebrate A-clade R2 proteins. Our work provides the first insights into A-clade R2 protein structure during gene insertion and may enable future improvement and adaptation of R2-based systems for precise transgene insertion.
- California Institute for Quantitative Biosciences (QB3), Berkeley, CA, USA.
Organizational Affiliation: