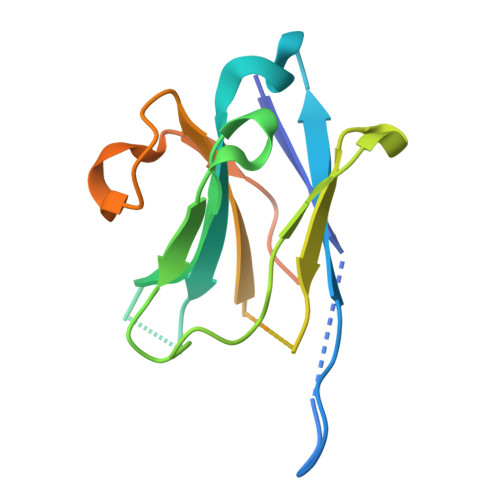
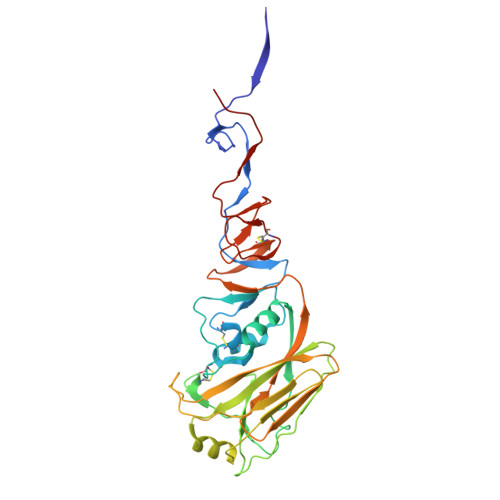
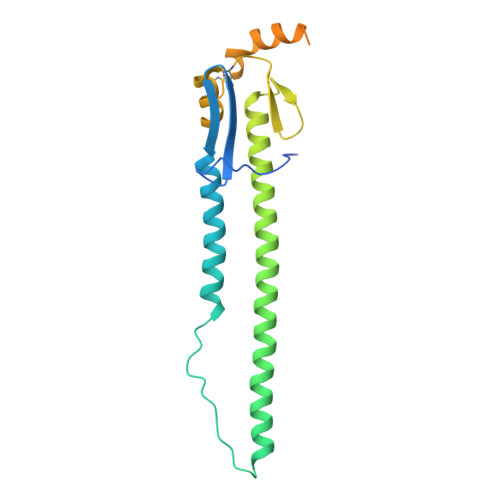
Atomically accurate de novo design of antibodies with RFdiffusion.
Bennett, N.R., Watson, J.L., Ragotte, R.J., Borst, A.J., See, D.L., Weidle, C., Biswas, R., Yu, Y., Shrock, E.L., Ault, R., Leung, P.J.Y., Huang, B., Goreshnik, I., Tam, J., Carr, K.D., Singer, B., Criswell, C., Wicky, B.I.M., Vafeados, D., Garcia Sanchez, M., Kim, H.M., Vazquez Torres, S., Chan, S., Sun, S.M., Spear, T.T., Sun, Y., O'Reilly, K., Maris, J.M., Sgourakis, N.G., Melnyk, R.A., Liu, C.C., Baker, D.(2026) Nature 649: 183-193
- PubMed: 41193805
- DOI: https://doi.org/10.1038/s41586-025-09721-5
- Primary Citation of Related Structures:
9NFU, 9NH7 - PubMed Abstract:
Despite the central role of antibodies in modern medicine, no method currently exists to design novel, epitope-specific antibodies entirely in silico. Instead, antibody discovery currently relies on immunization, random library screening or the isolation of antibodies directly from patients 1 . Here we demonstrate that combining computational protein design using a fine-tuned RFdiffusion 2 network with yeast display screening enables the de novo generation of antibody variable heavy chains (VHHs), single-chain variable fragments (scFvs) and full antibodies that bind to user-specified epitopes with atomic-level precision. We experimentally characterize VHH binders to four disease-relevant epitopes. Cryo-electron microscopy confirms the binding pose of designed VHHs targeting influenza haemagglutinin and Clostridium difficile toxin B (TcdB). A high-resolution structure of the influenza-targeting VHH confirms atomic accuracy of the designed complementarity-determining regions (CDRs). Although initial computational designs exhibit modest affinity (tens to hundreds of nanomolar K d ), affinity maturation using OrthoRep 3 enables production of single-digit nanomolar binders that maintain the intended epitope selectivity. We further demonstrate the de novo design of scFvs to TcdB and a PHOX2B peptide-MHC complex by combining designed heavy-chain and light-chain CDRs. Cryo-electron microscopy confirms the binding pose for two distinct TcdB scFvs, with high-resolution data for one design verifying the atomically accurate design of the conformations of all six CDR loops. Our approach establishes a framework for the computational design, screening and characterization of fully de novo antibodies with atomic-level precision in both structure and epitope targeting.
- Department of Biochemistry, University of Washington, Seattle, WA, USA.
Organizational Affiliation: