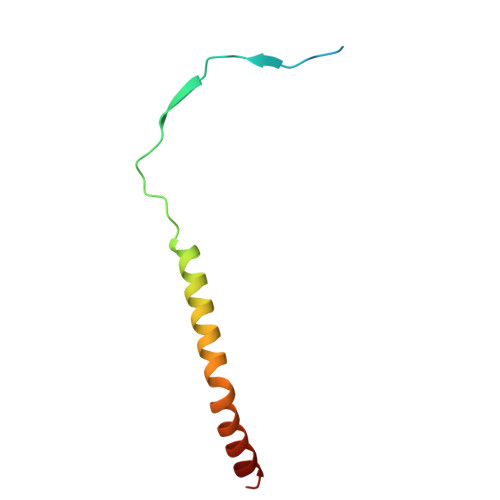
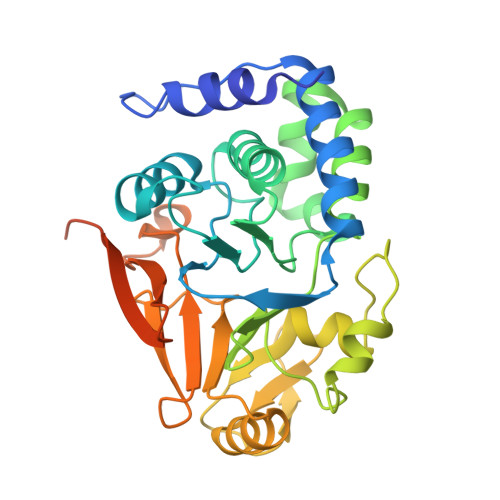
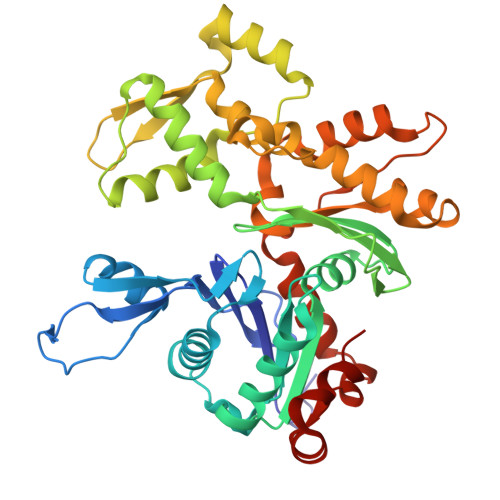
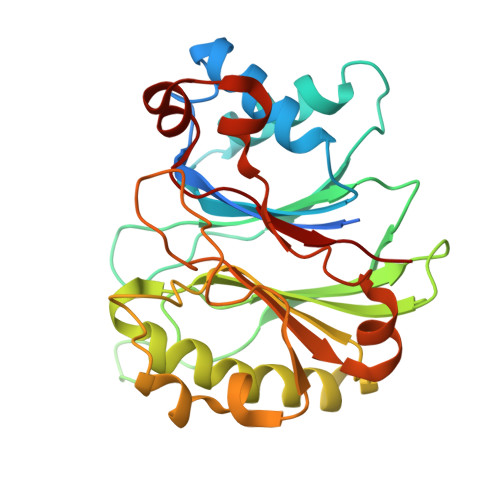
Harnessing the Evolution of Proteostasis Networks to Reverse Cognitive Dysfunction.
Reineke, L.C., Zhu, P.J., Dalwadi, U., Dooling, S.W., Liu, Y., Wang, I.C., Young-Baird, S., Okoh, J., Kuncha, S.K., Zhou, H., Kannan, A., Park, H., Debeaubien, N.A., Croll, T., Lee, D.J., Arthur, C., Dever, T.E., Walter, P., Chen, J., Frost, A., Costa-Mattioli, M.(2025) bioRxiv
- PubMed: 40568171
- DOI: https://doi.org/10.1101/2025.02.28.640897
- Primary Citation of Related Structures:
9NB9 - PubMed Abstract:
The integrated stress response (ISR) is a highly conserved network essential for maintaining cellular homeostasis and cognitive function. Here, we investigated how persistent ISR activation impacts cognitive performance, primarily focusing on a PPP1R15B R658C genetic variant associated with intellectual disability. By generating a novel mouse model that mimics this human condition, we revealed that this variant destabilizes the PPP1R15B•PP1 phosphatase complex, resulting in chronic ISR activation, impaired protein synthesis, and deficits in long-term memory. Importantly, we found that the cognitive and synaptic deficits in Ppp1r15b R658C mice are directly due to ISR activation. Leveraging insights from evolutionary biology, we characterized DP71L, a viral orthologue of PPP1R15B, through detailed molecular and structural analyses, uncovering its mechanism of action as a potent pan-ISR inhibitor. Remarkably, we found that DP71L not only buffers cognitive decline associated with a wide array of conditions-including Down syndrome, Alzheimer's disease and aging-but also enhances long-term synaptic plasticity and memory in healthy mice. These findings highlight the promise of utilizing evolutionary insight to inform innovative therapeutic strategies.