Discovery of TNG-6132, a potent, selective, and orally bioavailable USP1 inhibitor.
Throner, S., Feng, T., Andersen, J.N., Bandi, M., Engel, J.L., Gong, S., Gotur, D., Gu, L., Huang, A., Lazarides, K., Maxwell, J.P., McCarren, P., McMillan, B.J., Pham, T.V., Simoneau, A., Tsai, A., Whittington, D.A., Wilker, E., Zhang, M., Zhang, W.(2025) Bioorg Med Chem Lett 124: 130262-130262
- PubMed: 40315934
- DOI: https://doi.org/10.1016/j.bmcl.2025.130262
- Primary Citation of Related Structures:
9N9Y - PubMed Abstract:
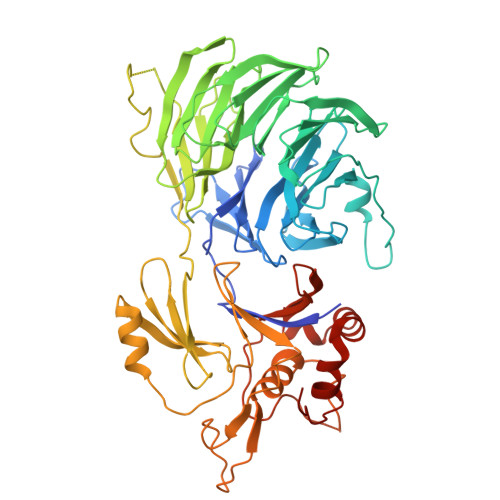
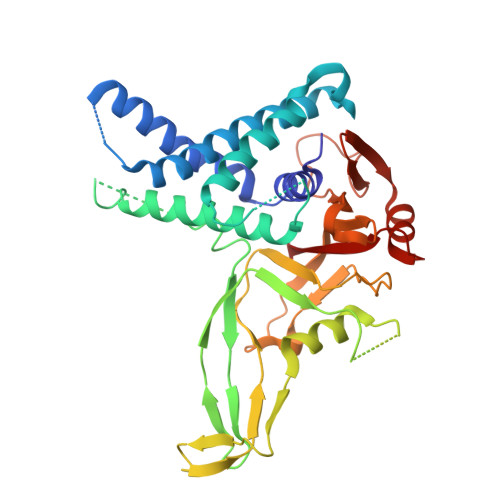
USP1 (ubiquitin-specific peptidase 1) is a deubiquitinating enzyme that has been identified as essential in BRCA1/2 mutant cells and implicated in the DNA damage response. Inhibition of USP1 by small molecule inhibitors disrupts DNA repair and replication and is being pursued as a potential anticancer therapeutic in BRCA1/2 mutant cancers. We report the discovery of an in vitro and in vivo USP1 inhibitor tool compound TNG-6132 (18), a reversible, allosteric inhibitor of USP1, which strongly inhibits USP1 enzymatic activity. This inhibitory effect translates into in vitro cellular viability defects in a BRCA1-mutant breast cancer cell line, as well as an in vivo pharmacodynamic (PD) response and tumor growth suppression in a mouse xenograft efficacy model. Additionally, we report an X-ray co-crystal structure of TNG-6132 (18) bound in the USP1-UAF1 complex, a result that furthered our understanding of the role played by key elements of the pharmacophore of this chemotype as well as its mechanism of inhibition of USP1.
- Tango Therapeutics, 201 Brookline Ave, Boston, MA 02215, United States.
Organizational Affiliation: