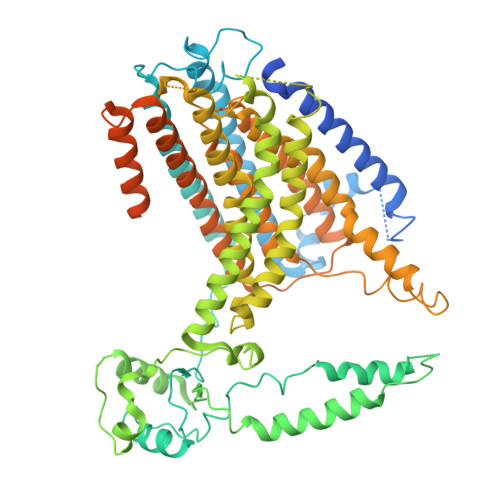
Structural and functional basis of mechanosensitive TMEM63 channelopathies.
Zheng, W., Lowry, A.J., Smith, H.E., Xie, J., Rawson, S., Wang, C., Ou, J., Sotomayor, M., Fu, T.M., Yang, H., Holt, J.R.(2025) Neuron 113: 2474
- PubMed: 40480214
- DOI: https://doi.org/10.1016/j.neuron.2025.05.009
- Primary Citation of Related Structures:
9N93, 9N95 - PubMed Abstract:
TMEM63A, -B, and -C constitute a mammalian family of mechanosensitive ion channels that are mutated in neurodevelopmental disorders. The molecular mechanisms underlying TMEM63 activation by force and the impact of disease-associated mutations have not been clarified. Here, we elucidate the structural and functional bases of a prevalent TMEM63B mutation p.V44M. We first found that TMEM63B p.V44M and the homologous TMEM63A p.V53M are gain-of-function mutations that do not enhance channel activity but instead evoke constitutive lipid scramblase activity. We then solved TMEM63A p.V53M mutant structures in both closed and lipid-open states, which revealed major rearrangements of pore-lining helices, creating a lateral cleft across the membrane. Simulation studies revealed lipid scrambling through this cleft. The structural rearrangements were triggered by disruption of a surface-proximal hydrophobic latch, a putative force-sensing module that includes a cluster of disease mutation sites. Our findings provide mechanistic insight into TMEM63 channelopathies and suggest a possible force-sensing mechanism.
- Departments of Otolaryngology & Neurology, Boston Children's Hospital and Harvard Medical School, Boston, MA 02115, USA; Department of Neuroscience, University of Wisconsin-Madison, Madison, WI 53705, USA. Electronic address: wzheng86@wisc.edu.
Organizational Affiliation: