A Bivalent Molecular Glue Linking Lysine Acetyltransferases to Oncogene-induced Cell Death.
Nix, M.N., Gourisankar, S., Sarott, R.C., Dwyer, B.G., Nettles, S.A., Martinez, M.M., Abuzaid, H., Yang, H., Wang, Y., Simanauskaite, J.M., Romero, B.A., Jones, H.M., Krokhotin, A., Lowensohn, T.N., Chen, L., Low, C., Davis, M.M., Fernandez, D., Zhang, T., Green, M.R., Hinshaw, S.M., Gray, N.S., Crabtree, G.R.(2025) bioRxiv
- PubMed: 40166243
- DOI: https://doi.org/10.1101/2025.03.14.643404
- Primary Citation of Related Structures:
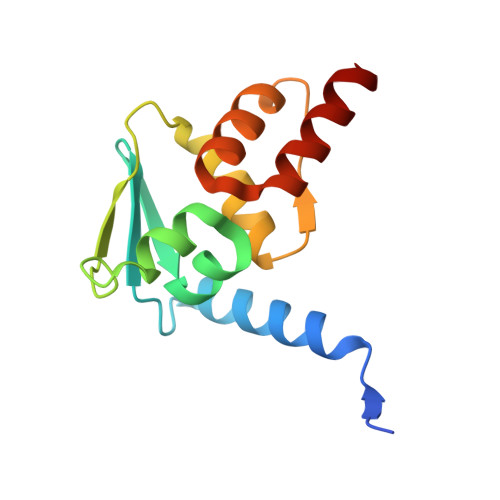
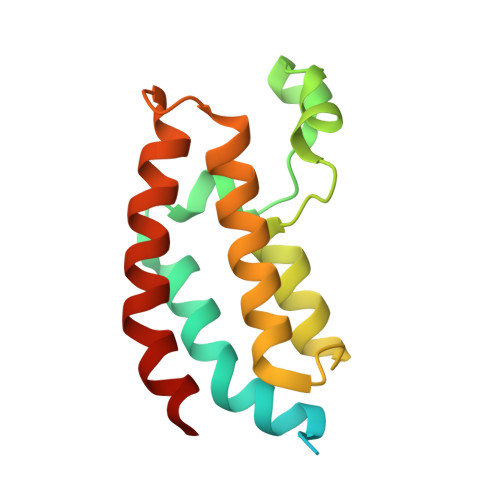
9MZA - PubMed Abstract:
Developing cancer therapies that induce robust death of the malignant cell is critical to prevent relapse. Highly effective strategies, such as immunotherapy, exemplify this observation. Here we provide the structural and molecular underpinnings for an approach that leverages chemical induced proximity to produce specific cell killing of diffuse large B cell lymphoma, the most common non-Hodgkin's lymphoma. We develop KAT-TCIPs (lysine acetyltransferase transcriptional/epigenetic chemical inducers of proximity) that redirect p300 and CBP to activate programmed cell death genes normally repressed by the oncogenic driver, BCL6. Acute treatment rapidly reprograms the epigenome to initiate apoptosis and repress c-MYC. The crystal structure of the chemically induced p300-BCL6 complex reveals how chance interactions between the two proteins can be systematically exploited to produce the exquisite potency and selectivity of KAT-TCIPs. Thus, the malignant function of an oncogenic driver can be co-opted to activate robust cell death, with implications for precision epigenetic therapies.