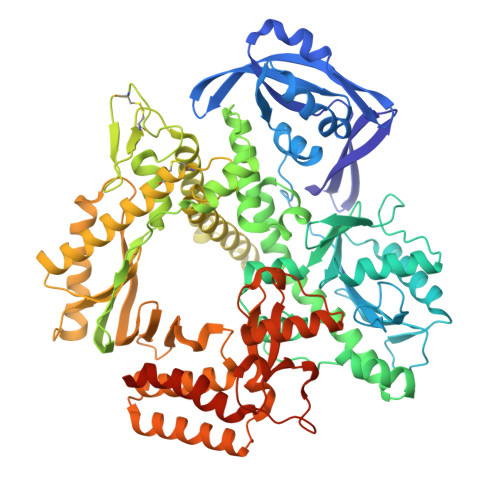
Rapid evolution of a highly efficient RNA polymerase by homologous recombination.
Medina, E.L., Maola, V.A., Hajjar, M., Ko, G.K., Ho, E.J., Horton, A.R., Chim, N., Chaput, J.C.(2026) Nat Chem Biol
- PubMed: 41501182
- DOI: https://doi.org/10.1038/s41589-025-02124-7
- Primary Citation of Related Structures:
9MYE - PubMed Abstract:
Engineering DNA polymerases to efficiently synthesize artificial or noncognate nucleic acids remains an essential challenge in synthetic biology. Here we describe an evolutionary campaign designed to convert a family of highly selective DNA polymerases into an unnatural homolog with strong RNA synthesis activity. Starting from a homologous recombination library, a short evolutionary path was achieved using a single-cell droplet-based microfluidic selection strategy to produce C28, a newly engineered polymerase that can synthesize RNA with a rate of ~3 nt s -1 and of >99% fidelity. C28 is capable of long-range RNA synthesis, reverse transcription and chimeric DNA-RNA amplification using the PCR. Despite strong discrimination against other genetic systems, C28 readily accepts several 2'F and base-modified RNA analogs. Together, these findings highlight the power of directed evolution as an approach for reprogramming DNA polymerases with activities that could help drive future applications in biotechnology and medicine.
- Department of Pharmaceutical Sciences, University of California, Irvine, Irvine, CA, USA.
Organizational Affiliation: