Beyond the beta-alpha-beta Fold: Characterization of a SnoaL Domain in the Tautomerase Superfamily.
Melkonian, T.R., Vuksanovic, N., Person, M.D., Chen, T.Y., Chang, W.C., Allen, K.N., Whitman, C.P.(2025) Biochemistry 64: 1950-1962
- PubMed: 40231412
- DOI: https://doi.org/10.1021/acs.biochem.5c00051
- Primary Citation of Related Structures:
9MUA, 9MUP - PubMed Abstract:
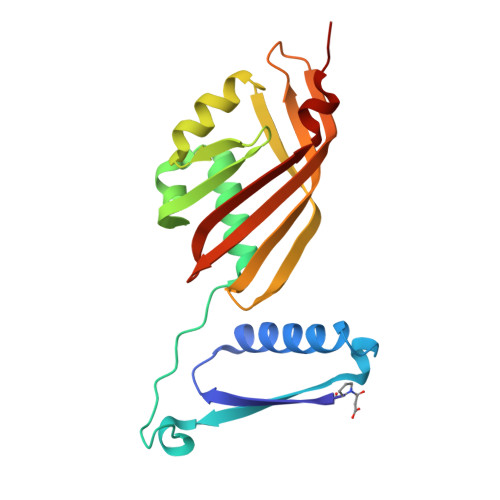
Tautomerase superfamily (TSF) members are constructed from a single β-α-β unit or two consecutively joined β-α-β units, and most have a catalytic Pro1. This pattern prevails throughout the superfamily consisting of more than 11,000 members where homo- or heterohexamers are localized in the 4-oxalocrotonate tautomerase (4OT)-like subgroup and trimers are found in the other four subgroups except for a small subset of 4OT trimers, symmetric and asymmetric, that are found in the 4OT-like subgroup. During a sequence similarity network (SSN) update, a small cluster of sequences (117 sequences) was discovered in the 4OT-like subgroup that begins with Pro1. These sequences consist of a 4OT-like domain fused to a SnoaL domain at the C-terminus (except for one), as annotated in the UniProt database. The Pseudooceanicola atlanticus one (designated "4OT-SnoaL") was chosen for kinetic, mechanistic, and crystallographic analysis. 4OT-SnoaL did not display detectable activity with known TSF substrates, suggesting a new activity. A genome neighborhood diagram (GND) places 4OT-SnoaL in an operon for a hydantoin degradation/utilization pathway. Treatment of 4OT-SnoaL with 3-bromopropiolate results in covalent modification of Pro1 by a 3-oxopropanoate adduct. Crystallographic analysis of the apo and modified enzymes shows that the 4OT domain is a hexamer of six identical subunits (a trimer of dimers), where each dimer consists of two β-α-β building blocks. Each C-terminus is attached to a SnoaL-like domain that displays a distorted α + β-barrel. The motif is a new one in the TSF and adds structural diversity to the TSF by using a SnoaL-like domain.
- Division of Chemical Biology and Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin, Austin, Texas 78712, United States.
Organizational Affiliation: