Engineering antibody-drug conjugates targeting an adhesion GPCR, CD97.
Hattori, T., Wang, M., Corrado, A.D., Gross, S., Fang, M., Bang, I., Roy, N., Berezniuk, I., Donaldson, H., Groff, K., Ravn-Boess, N., Koide, A., Placantonakis, D.G., Park, C.Y., Koide, S.(2025) Proc Natl Acad Sci U S A 122: e2516627122-e2516627122
- PubMed: 41026810
- DOI: https://doi.org/10.1073/pnas.2516627122
- Primary Citation of Related Structures:
9MQR - PubMed Abstract:
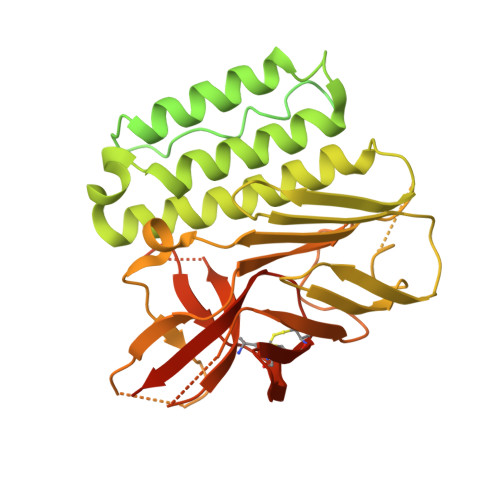
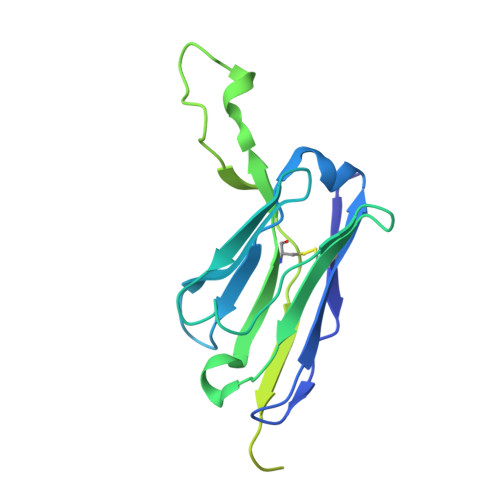
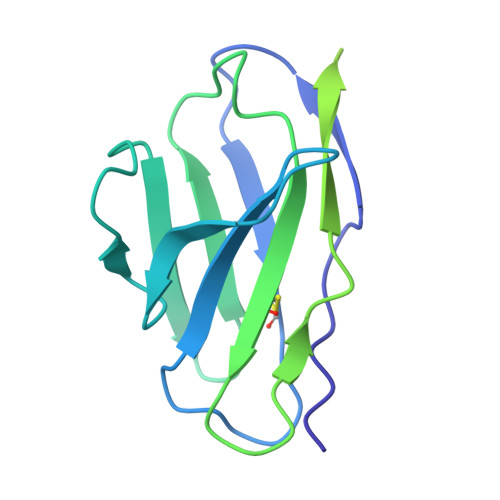
Adhesion G protein-coupled receptors (aGPCRs) are key cell-adhesion molecules involved in many cellular functions and contribute to human diseases, including cancer. aGPCRs are characterized by large extracellular regions that could serve as readily accessible antigens. However, the potential of aGPCRs as targets for biologic therapeutics has not been extensively explored. CD97, also known as ADGRE5, is an aGPCR that is upregulated in various cancer types, including acute myeloid leukemia (AML) and glioblastoma (GBM), and their respective cancer stem cells. Here, we developed antibody-drug conjugates (ADCs) targeting CD97 and assessed their efficacy against AML and GBM cells. We generated a panel of synthetic human antibodies targeting distinct epitopes of CD97, from which we identified an antibody that was efficiently internalized. This antibody binds to all isoforms of human CD97 but not to its close homolog, EMR2. Structure determination by single-particle cryo-electron microscopy revealed that this antibody targets the CD97 GPCR autoproteolysis-inducing (GAIN) domain, whose presence is conserved in aGPCRs, through an unconventional binding mode where it extensively utilizes the light chain framework for antigen recognition. Screening of conjugation methods and payloads resulted in a stable ADC that effectively killed AML and GBM cell lines, as well as patient-derived GBM stem cells, with minimal cytotoxicity against peripheral blood mononuclear cells from healthy donors. Our study demonstrates the therapeutic potential of targeting CD97, as well as the aGPCR GAIN domain in general, and uncovers a previously unrecognized surface that an antibody can utilize for antigen recognition.
- Perlmutter Cancer Center, New York University Langone Health, New York, NY 10016.
Organizational Affiliation: