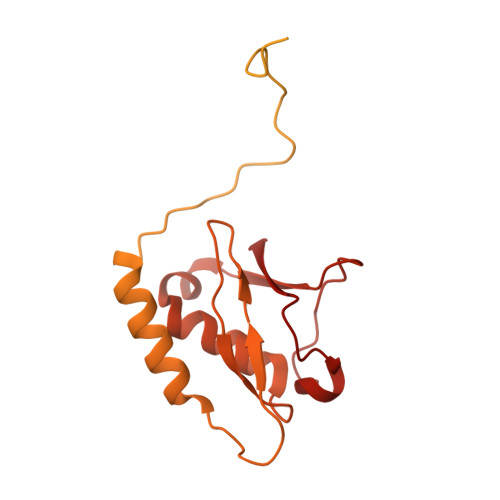
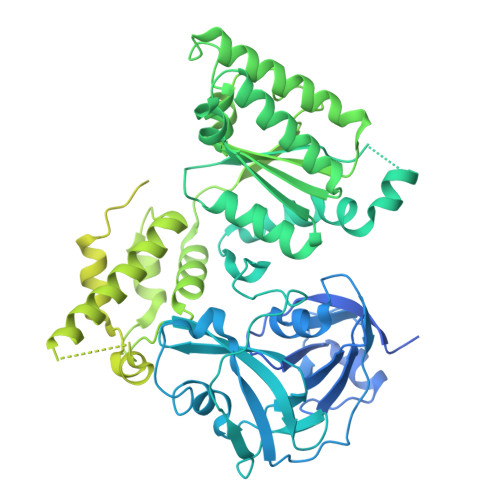
Structural basis of VCP-VCPIP1-p47 ternary complex in Golgi maintenance.
Shah, B., Hunkeler, M., Bratt, A., Yue, H., Jaen Maisonet, I., Fischer, E.S., Buhrlage, S.J.(2025) Nat Commun 16: 8025-8025
- PubMed: 40877265
- DOI: https://doi.org/10.1038/s41467-025-63161-3
- Primary Citation of Related Structures:
9MPQ, 9MPR, 9MPS, 9MPT, 9MPU, 9MPV - PubMed Abstract:
VCP/p97 regulates a wide range of cellular processes, including post-mitotic Golgi reassembly. In this context, VCP is assisted by p47, an adapter protein, and VCPIP1, a deubiquitylase (DUB). However, how they organize into a functional ternary complex to promote Golgi assembly remains unknown. Here, we use cryo-EM to characterize both VCP-VCPIP1 and VCP-VCPIP1-p47 complexes. We show that VCPIP1 engages VCP through two interfaces: one involving the N-domain of VCP and the UBX domain of VCPIP1, and the other involving the VCP D2 domains and a region of VCPIP1 we refer to as VCPID. The p47 UBX domain competitively binds to the VCP N-domain, while not affecting VCPID binding. We show that VCPID is critical for VCP-mediated enhancement of DUB activity and proper Golgi assembly. The ternary structure along with biochemical and cellular data provides new insights into the complex interplay of VCP with its co-factors.
- Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, USA.
Organizational Affiliation: