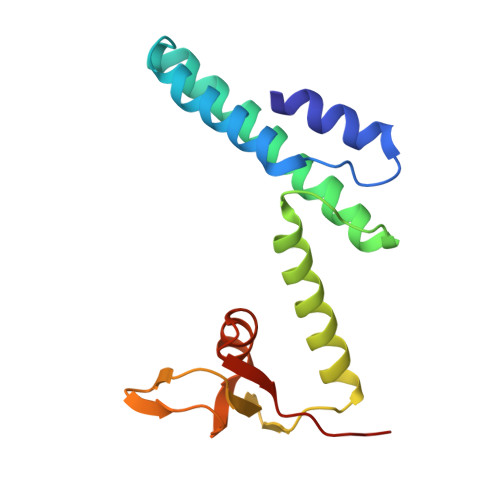
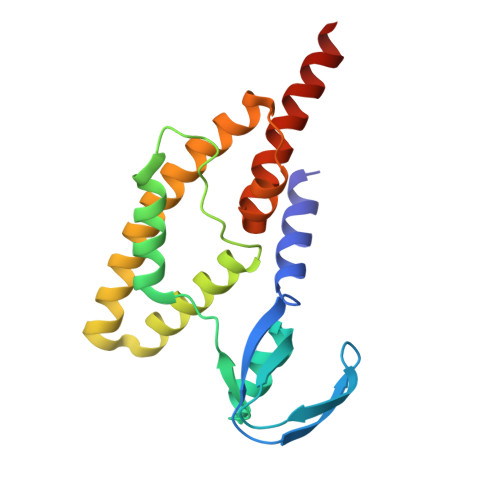
Structure of the Saccharolobus solfataricus GINS tetramer.
Shankar, S., Enemark, E.J.(2025) Acta Crystallogr F Struct Biol Commun 81: 207-215
- PubMed: 40235367
- DOI: https://doi.org/10.1107/S2053230X25003085
- Primary Citation of Related Structures:
9MOJ - PubMed Abstract:
DNA replication is tightly regulated to ensure genomic stability and prevent several diseases, including cancers. Eukaryotes and archaea partly achieve this regulation by strictly controlling the activation of hexameric minichromosome maintenance (MCM) helicase rings that unwind DNA during its replication. In eukaryotes, MCM activation critically relies on the sequential recruitment of the essential factors Cdc45 and a tetrameric GINS complex at the onset of the S-phase to generate a larger CMG complex. We present the crystal structure of the tetrameric GINS complex from the archaeal organism Saccharolobus solfataricus (Sso) to reveal a core structure that is highly similar to the previously determined GINS core structures of other eukaryotes and archaea. Using molecular modeling, we illustrate that a subdomain of SsoGINS would need to move to accommodate known interactions of the archaeal GINS complex and to generate a SsoCMG complex analogous to that of eukaryotes.
- Department of Biochemistry and Molecular Biology, University of Arkansas for Medical Sciences, 4301 West Markham Street, Slot 516, Little Rock, AR 72205, USA.
Organizational Affiliation: