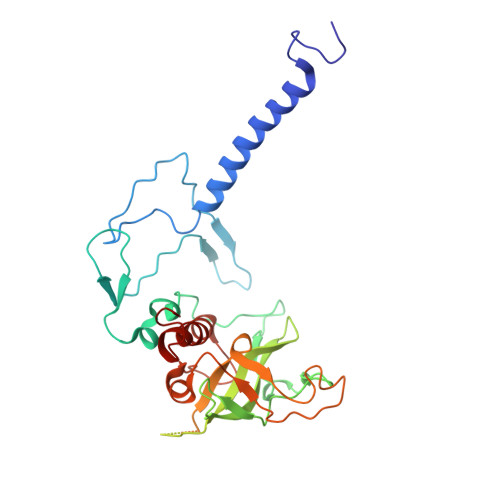
Structure and Biochemistry of the LH1-RC Photocomplex from the Halophilic Purple Bacterium, Rhodothalassium salexigens .
Tani, K., Kanno, R., Inami, M., Ooya, T., Matsushita, R., Inada, K., Takenaka, S., Takaichi, S., Purba, E.R., Hall, M., Mochizuki, T., Yu, L.J., Mizoguchi, A., Humbel, B.M., Madigan, M.T., Kimura, Y., Wang-Otomo, Z.Y.(2025) Biochemistry 64: 3070-3080
- PubMed: 40551662
- DOI: https://doi.org/10.1021/acs.biochem.5c00181
- Primary Citation of Related Structures:
9M8M - PubMed Abstract:
Rhodothalassium (Rts.) salexigens is a moderately halophilic purple nonsulfur bacterium whose unique cell wall composition and phylogeny are distinct from those of all other purple phototrophs. Here we present a cryo-EM structure and biochemical analysis of the light-harvesting 1-reaction center (LH1-RC) complex from Rts. salexigens at 2.29 Å resolution. The LH1 complex forms a closed ring structure with 16 αβ-polypeptides surrounding the RC and contains 16 phosphatidylglycerols regularly positioned between the β-polypeptides. Extensive interactions were observed between the C-terminal domains of LH1 α-and β-polypeptides and between the regularly arranged phosphatidylglycerols and β-polypeptides, supporting the hypothesis that LH1 C-terminal interactions define the post-translational truncation sites of αβ-polypeptides in phototrophic purple bacteria. Multiple insertions were identified in the membrane-extruded RC cytochrome- and H-subunits of Rts. salexigens . Insertions in the periplasm-exposed cytochrome subunit contain high proportions of Gly, Asp, and Glu, contributing to an overall negatively charged surface of this subunit. The cytoplasm-exposed H-subunit contained an unusually long 57-residue insert rich in Pro and Ala that was invisible in the cryo-EM density map, indicating its highly flexible nature. The extensive Pro-Ala repetitive motifs in this insertion points to a regulatory role in assemblies of the RC and LH1-RC complexes. The structural features of Rts. salexigens LH1-RC are also discussed in relation to differences in the physiological environment between the periplasmic and cytoplasmic sides of membranes in halophiles, necessary for maintaining cellular activities under the high ionic strength conditions of hypersaline environments.
- Center for Computational Sciences, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8577, Japan.
Organizational Affiliation: