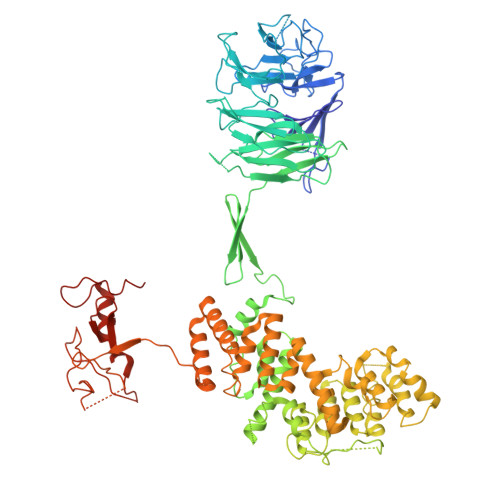
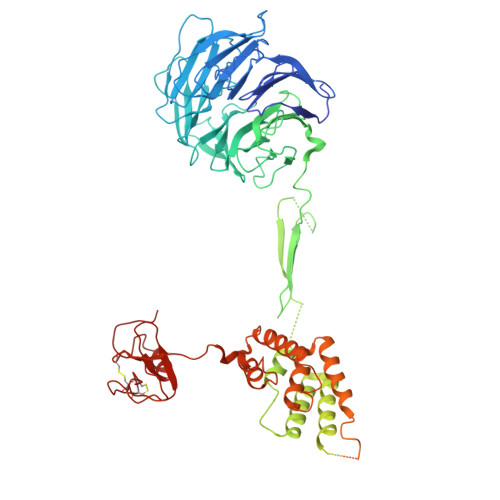
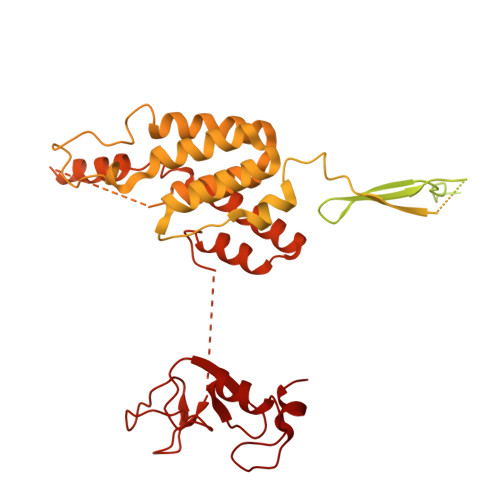
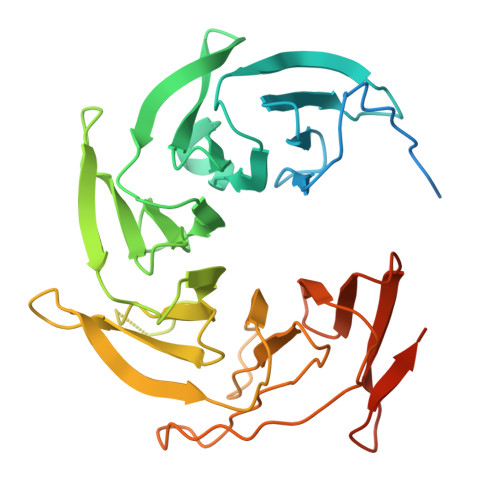
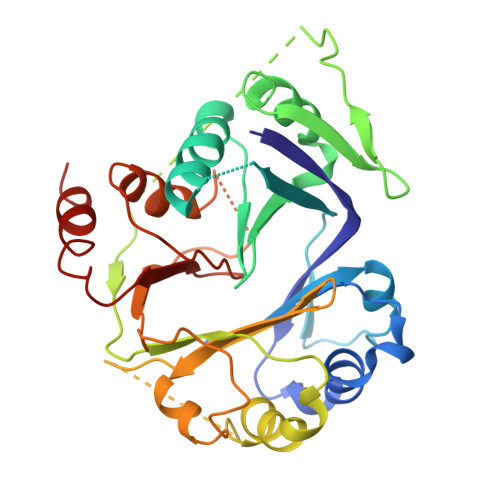
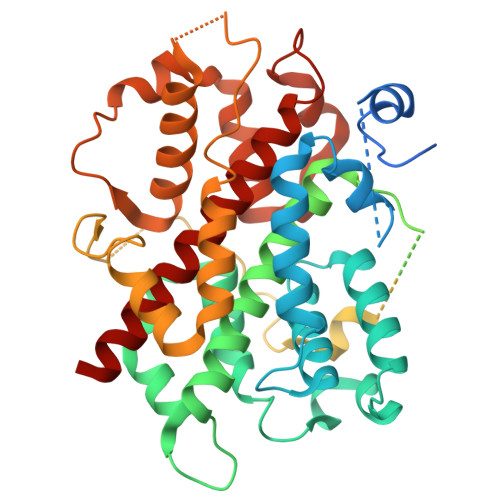
Cryo-EM structures of amino acid sensors bound to the human GATOR2 complex.
Su, M.Y., Teng, F., Wang, S., Mai, X., Zeng, H., Li, J., Song, X., Wang, X., Stjepanovic, G.(2025) Cell Rep 44: 116088-116088
- PubMed: 40742811
- DOI: https://doi.org/10.1016/j.celrep.2025.116088
- Primary Citation of Related Structures:
9LVJ, 9LVK, 9LWF - PubMed Abstract:
Mammalian cells regulate growth by integrating environmental cues through the mammalian target of rapamycin complex 1 (mTORC1) signaling pathway. The human GATOR2 complex, comprising WDR59, WDR24, Mios, Sec13, and Seh1l, is key to mTORC1 regulation. Under amino acid deprivation, GATOR2 is inhibited through interactions with cytosolic leucine sensor Sestrin2 and arginine sensor cytosolic arginine sensor for mTORC1 subunit 1 (CASTOR1). Amino acid abundance relieves this inhibition, allowing GATOR2 to antagonize the repressor GATOR1. Despite its importance, GATOR2's inhibition mechanisms were unclear. Here, we present cryo-electron microscopy (cryo-EM) structures of GATOR2 in three inhibitory states: CASTOR1 bound, Sestrin2 bound, and dual bound. CASTOR1 engages the Mios WD40 β-propellers, while Sestrin2 interacts with the WDR24-Seh1l subcomplex, inducing conformational movements. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) reveals dynamic motions in apo-GATOR2 and its complexes with amino acid sensors, as well as the effects of amino acid supplementation. These findings unravel the interactions between GATOR2 and amino acid sensors, providing a perspective on the regulation of the mTORC1 pathway by nutrient-sensing machinery.
- Department of Biochemistry, Key University Laboratory of Metabolism and Health of Guangdong, SUSTech Homeostatic Medicine Institute, School of Medicine, Southern University of Science and Technology, Shenzhen 518055, China; Institute for Biological Electron Microscopy, Southern University of Science and Technology, Shenzhen 518055, China. Electronic address: sumy@sustech.edu.cn.
Organizational Affiliation: