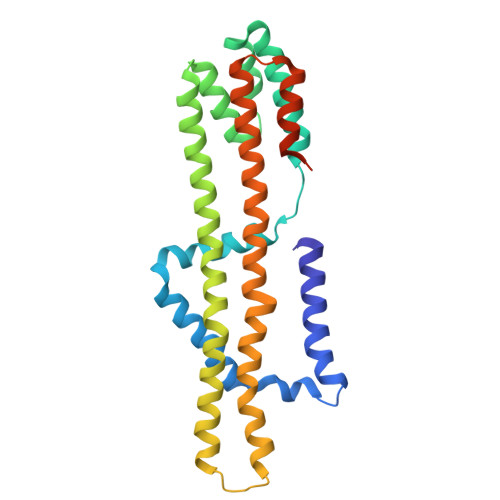
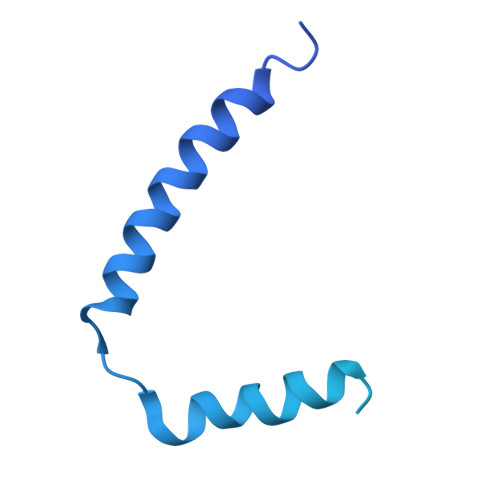
Cryo-EM Structure of the Flagellar Motor Complex from Paenibacillus sp. TCA20.
Onoe, S., Nishikino, T., Kinoshita, M., Takekawa, N., Minamino, T., Imada, K., Namba, K., Kishikawa, J.I., Kato, T.(2025) Biomolecules 15
- PubMed: 40149971
- DOI: https://doi.org/10.3390/biom15030435
- Primary Citation of Related Structures:
9LU9, 9LUB, 9LUC - PubMed Abstract:
The bacterial flagellum, a complex nanomachine composed of numerous proteins, is utilized by bacteria for swimming in various environments and plays a crucial role in their survival and infection. The flagellar motor is composed of a rotor and stator complexes, with each stator unit functioning as an ion channel that converts flow from outside of cell membrane into rotational motion. Paenibacillus sp. TCA20 was discovered in a hot spring, and a structural analysis was conducted on the stator complex using cryo-electron microscopy to elucidate its function. Two of the three structures (Classes 1 and 3) were found to have structural properties typical for other stator complexes. In contrast, in Class 2 structures, the pentamer ring of the A subunits forms a C-shape, with lauryl maltose neopentyl glycol (LMNG) bound to the periplasmic side of the interface between the A and B subunits. This interface is conserved in all stator complexes, suggesting that hydrophobic ligands and lipids can bind to this interface, a feature that could potentially be utilized in the development of novel antibiotics aimed at regulating cell motility and infection.
- Institute for Protein Research, Osaka University, Suita 565-0871, Osaka, Japan.
Organizational Affiliation: