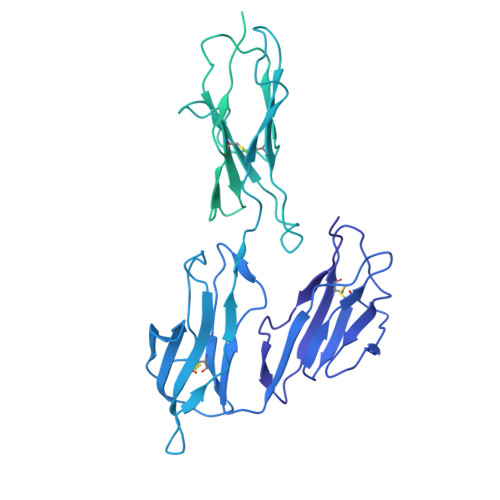
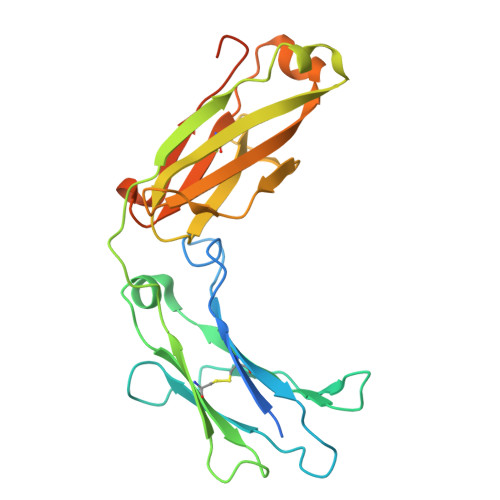
Human FcRL5 is an Fc receptor that simultaneously engages two IgGs.
Chen, S., Li, S., Zhang, Z., Xiao, J.(2026) Sci Adv 12: eaeb8865-eaeb8865
- PubMed: 41477863
- DOI: https://doi.org/10.1126/sciadv.aeb8865
- Primary Citation of Related Structures:
9LOC, 9LOD - PubMed Abstract:
The Fc receptors play crucial roles in initiating the effector functions of immunoglobulins. FcRL5 is a prominent target in B cell malignancies and has been implicated as a receptor for immunoglobulin G (IgG). However, the molecular mechanism remained unclear. Here, we demonstrate that human FcRL5, but not its mouse counterpart, is a bona fide IgG-Fc (Fcγ) receptor that uniquely requires the presence of two Fcγ molecules in close proximity to form a robust interaction. Cryo-electron microscopy reveals that FcRL5 optimally engages two Fcγ molecules positioned at a 60° angle, with its D1-D2 domains binding to the first Fcγ molecule, while its D3 domain arches over the second Fcγ. This distinctive binding capability enables FcRL5 to specifically recognize IgG immune complexes (ICs), with the binding strength correlating with IgG concentration in the ICs. In addition, we demonstrate that FcRL5 can internalize the IgG polymer and IC. These results shed light on FcRL5 function and reveal a unique Fcγ-FcγR binding mode governed by avidity.
- Joint Graduate Program of Peking-Tsinghua-NIBS, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing, P.R. China.
Organizational Affiliation: