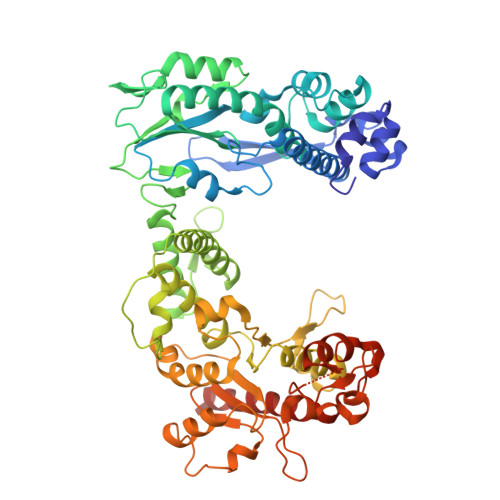
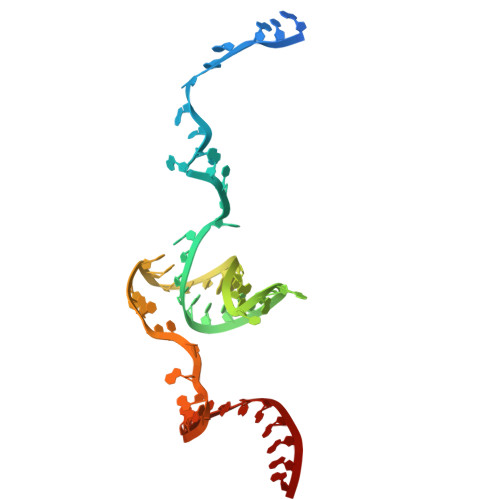
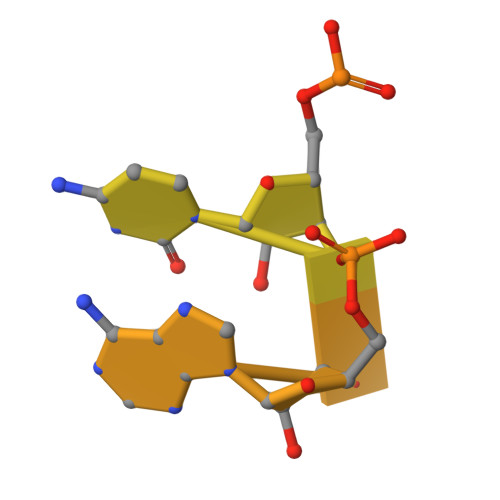
Structural basis of the RNA-mediated Retron-Eco2 oligomerization.
Wang, Y., Wang, C., Yin, Y., Cui, Y., Dai, Z., Liu, C., Chen, Y., Guan, Z., Zou, T.(2025) Cell Discov 11: 73-73
- PubMed: 40897710
- DOI: https://doi.org/10.1038/s41421-025-00823-y
- Primary Citation of Related Structures:
9LM3 - PubMed Abstract:
In the evolutionary arms race between bacteria and viruses, retrons have emerged as distinctive antiphage defense systems. Here, we elucidate the structure and function of Retron-Eco2, which comprises a non-coding RNA (ncRNA) that encodes multicopy single-stranded DNA (msDNA, a DNA‒RNA hybrid) and a fusion protein containing a reverse transcriptase (RT) domain and a topoisomerase-primase-like (Toprim) effector domain. The Eco2 msDNA and RT-Toprim fusion protein form a 1:1 stoichiometric nucleoprotein complex that further assembles into a trimer (msDNA:RT-Toprim ratio of 3:3) with a distinctive triangular configuration. The RNA portion of the msDNA in one protomer closely intertwines around the RT domain of an adjacent protomer, mediating the formation of this self-inhibitory assembly. Upon activation, the Toprim effector domain exhibits RNase activity, degrading RNA to arrest phage replication. We further reveal that phage mutants evading Eco2-mediated defense harbor mutations in the endonuclease IV-like protein DenB, underscoring DenB's critical role in triggering the activation of this system. Together, these findings provide key structural and functional insights into Retron-Eco2, laying the groundwork for harnessing its potential in biotechnology and synthetic biology applications.
- National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei, China.
Organizational Affiliation: