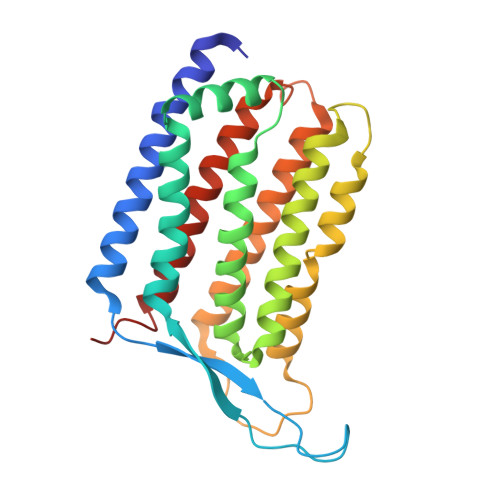
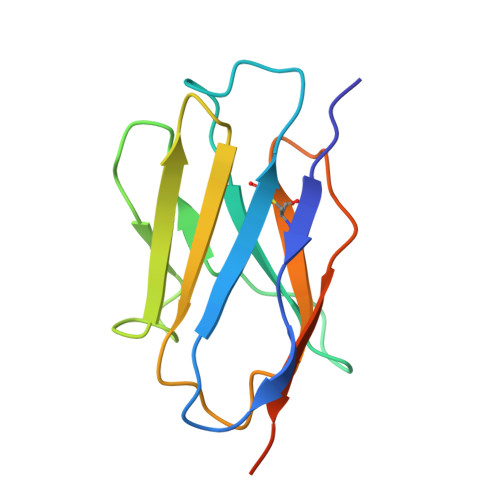
Cryo-EM structure of a nanobody-bound heliorhodopsin.
Xia, R., Sun, M., Lu, Y., Wang, N., Zhang, A., Guo, C., Xu, Z., Cai, X., He, Y.(2025) Biochem Biophys Res Commun 750: 151398-151398
- PubMed: 39889627
- DOI: https://doi.org/10.1016/j.bbrc.2025.151398
- Primary Citation of Related Structures:
9LJJ - PubMed Abstract:
Heliorhodopsins (HeRs) represent a distinct class of microbial rhodopsins (MRs) with an inverted membrane topology compared to other MRs. Previous structural studies have shown that HeRs lack a proton acceptor residue, and protons are never released from the protein. In this study, we present the cryo-electron microscopy (cryo-EM) structure of HeR bound to a nanobody. The structure reveals an acetate-like molecule in the Schiff base cavity (SBC) on the intracellular side of HeR under neutral condition. Structural comparisons and analyses suggest that the acetate molecule may function as a proton acceptor for the protonated retinal Schiff base (RSB) and act as a mediator for the intramolecular signaling transduction in HeR during light stimulation. These structural insights shed new light on the mechanism and function of HeR.
- HIT Center for Life Sciences, School of Life Science and Technology, Faculty of Life Sciences and Medicine, Harbin Institute of Technology, Harbin, 150001, China.
Organizational Affiliation: