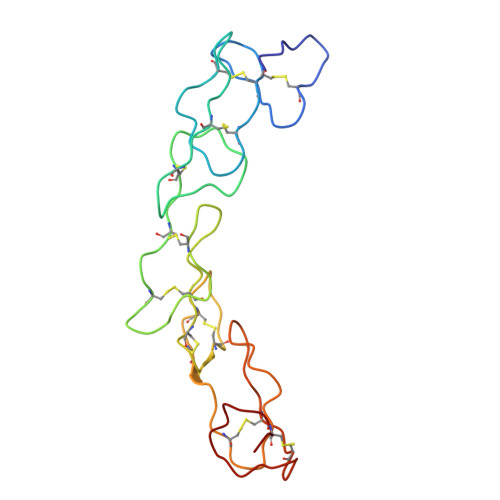
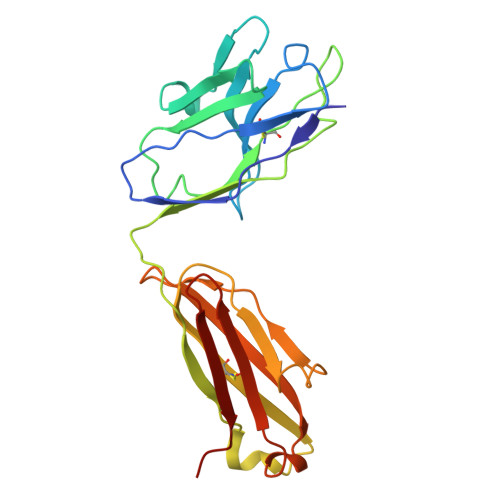
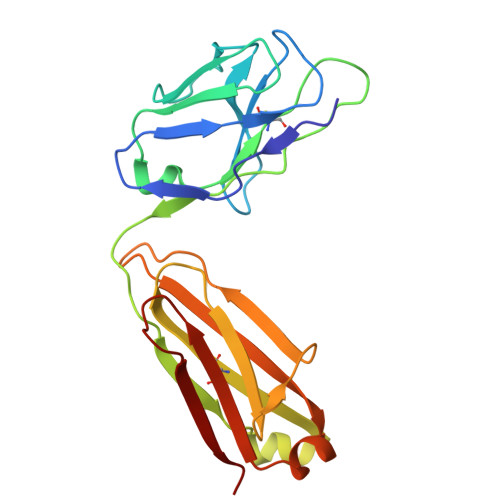
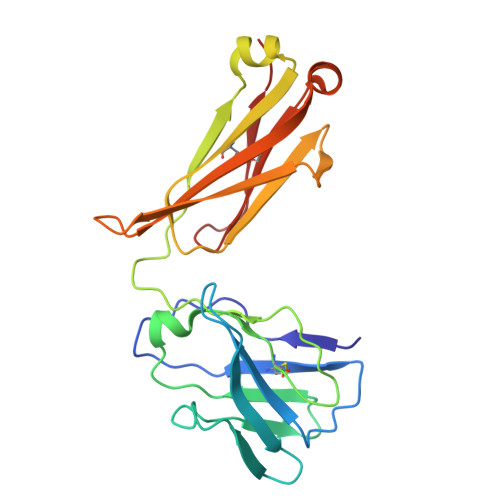
Conversion of an agonistic anti-TNFR2 biparatopic antibody into an antagonist by insertion of peptide linkers into the hinge region.
Otsuki, T., Matsumoto, S., Fujita, J., Miyata, T., Namba, K., Kanada, R., Okuno, Y., Kamada, H., Ohno, H., Akiba, H.(2025) J Biological Chem 301: 110548-110548
- PubMed: 40752574
- DOI: https://doi.org/10.1016/j.jbc.2025.110548
- Primary Citation of Related Structures:
9LFL - PubMed Abstract:
Biparatopic antibodies (BpAbs) bind two different antigen epitopes to form characteristic immunocomplexes. Many BpAbs have been developed for enhanced cross-linking to induce signal transduction or cell internalization, whereas few were reported with smaller immunocomplexes to suppress unwanted signaling. Here, we developed a strategy to induce 1:1 immunocomplex formation to maximize antagonistic function. Various peptide linkers were introduced into the hinge regions of IgG-like agonist BpAbs against tumor necrosis factor receptor 2. Loss of crosslinking activity was observed for one BpAb, allowing the conversion of its function from an agonist to an antagonist. However, cross-linking activity was retained for another agonist BpAb, which binds to a different epitope pair. In a combined analysis of cryo-electron microscopy and coarse-grained molecular dynamics simulations, effect of epitope combination on the stability of 1:1 complexes was observed. These results lead to an understanding of the mechanism and design of BpAbs to adopt a 1:1-binding mode.
- Graduate School of Pharmaceutical Sciences, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan.
Organizational Affiliation: