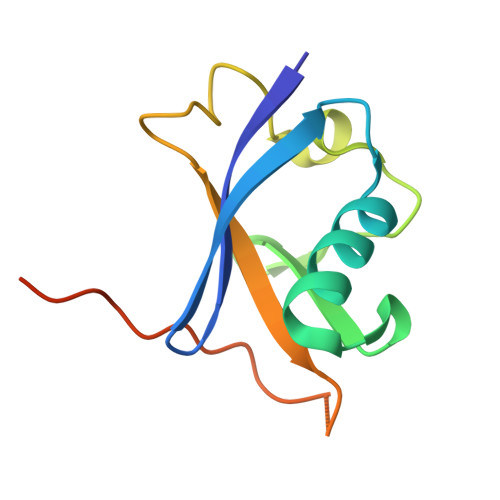
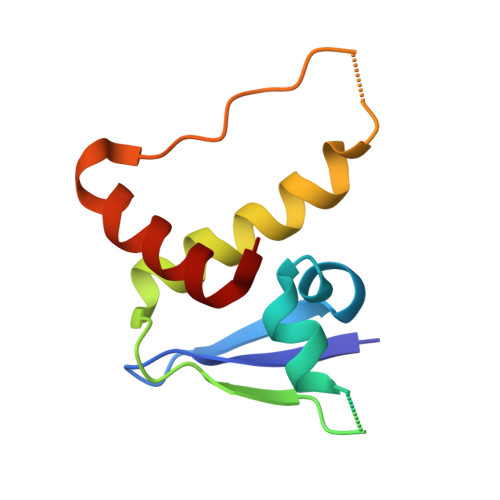
Structural analysis of EPOP BC-box binding to the elongin BC complex.
Kim, S., Yeo, H., Lee, B.I.(2025) Biochem Biophys Res Commun 759: 151691-151691
- PubMed: 40153999
- DOI: https://doi.org/10.1016/j.bbrc.2025.151691
- Primary Citation of Related Structures:
9LAF - PubMed Abstract:
The elongin BC complex (ELOBC) interacts with BC-box-containing proteins and plays a role in various cellular processes, including transcriptional regulation and ubiquitination. Elongin BC and polycomb repressive complex 2-associated protein (EPOP) contains a BC-box motif in its N-terminal region and influences cancer cell proliferation and differentiation. A previous study showed that a BC-box containing an EPOP-derived peptide suppresses cancer cell growth and induces apoptosis by disrupting the interaction between the ELOBC and its partner proteins. Here, we report the crystal structure of the EPOP BC-box peptide bound to the ELOBC and compare it with the structures of other BC-box-containing proteins in complex with the ELOBC. The overall structure of interactions between the BC-box and the ELOC was similar across different complexes, indicating a conserved binding mode. Our structural analysis revealed that the strictly conserved leucine residue (Leu40) within the BC-box of EPOP, which was previously suggested to be critical for interactions between the BC-box and the ELOC, was deeply embedded in the hydrophobic pocket of the ELOC protein. This study provided structural insights into BC-box-mediated protein-protein interactions and may serve as a fundamental resource for developing small molecules that modulate the interactions between ELOBC and BC-box-containing proteins.
- Research Institute, National Cancer Center, Goyang-si, Gyeonggi-do, 10408, Republic of Korea; Department of Cancer Biomedical Science, National Cancer Center Graduate School of Cancer Science and Policy, Goyang-si, Gyeonggi-do, 10408, Republic of Korea.
Organizational Affiliation: