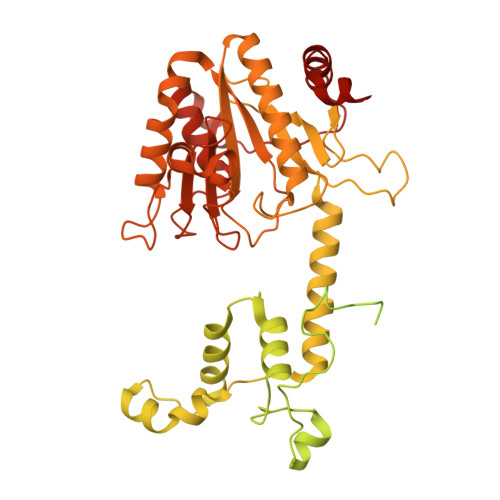
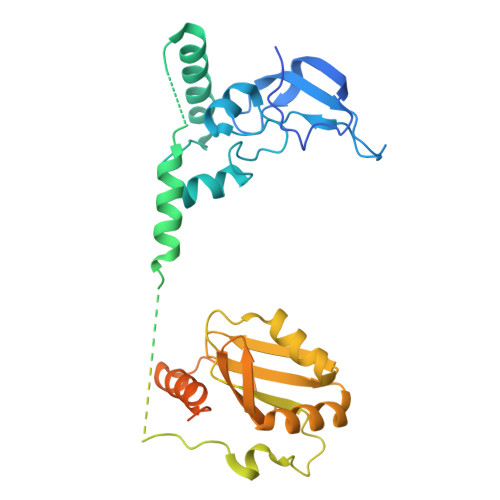
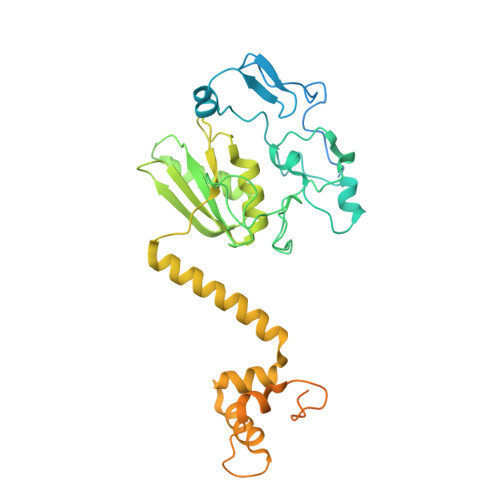
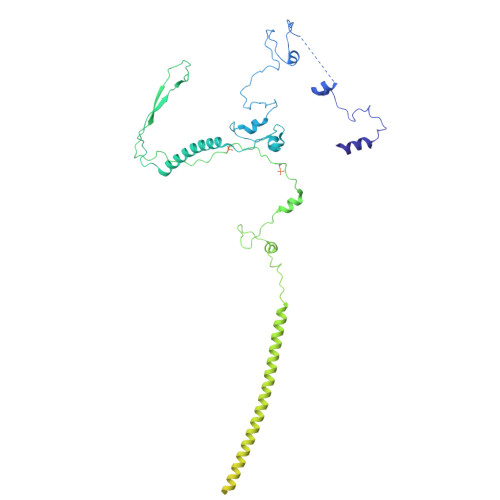
Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Li, Y., Fischer, P., Wang, M., Zhou, Q., Song, A., Yuan, R., Meng, W., Chen, F.X., Luhrmann, R., Lau, B., Hurt, E., Cheng, J.(2025) Cell Res 35: 296-308
- PubMed: 40016598
- DOI: https://doi.org/10.1038/s41422-025-01084-w
- Primary Citation of Related Structures:
9L5R, 9L5S, 9L5T - PubMed Abstract:
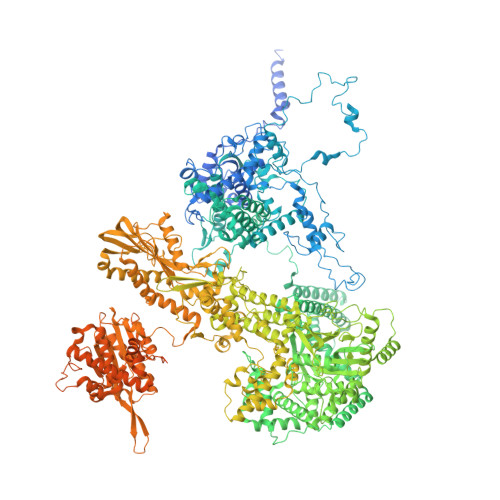
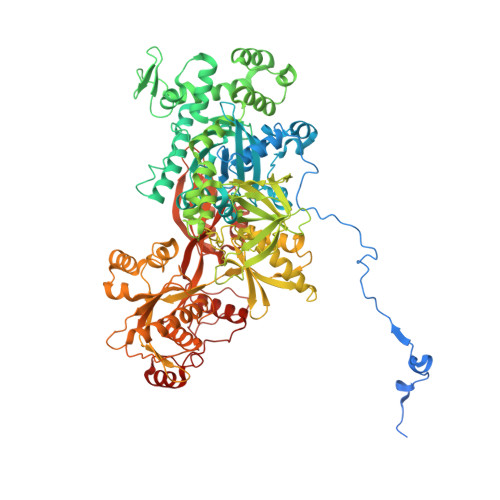
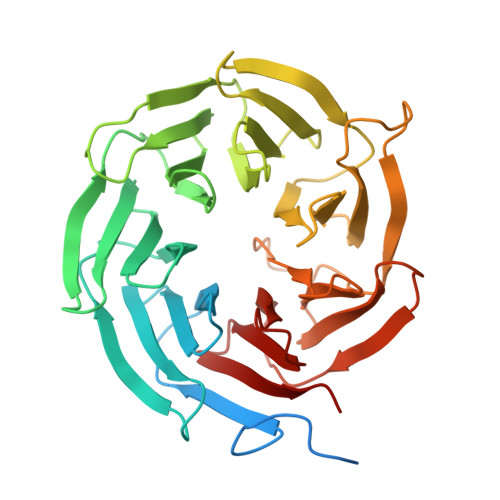
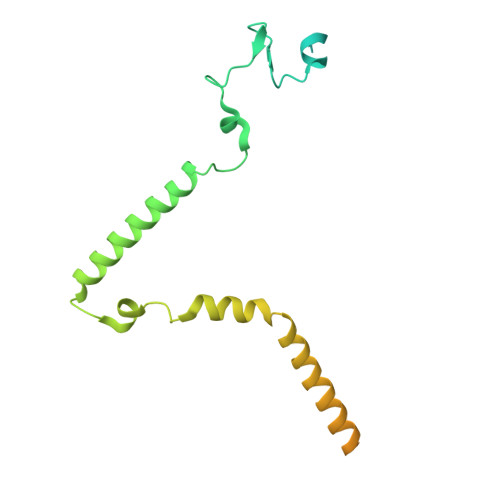
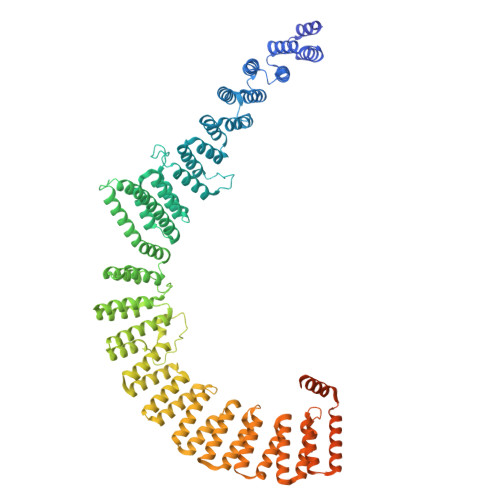
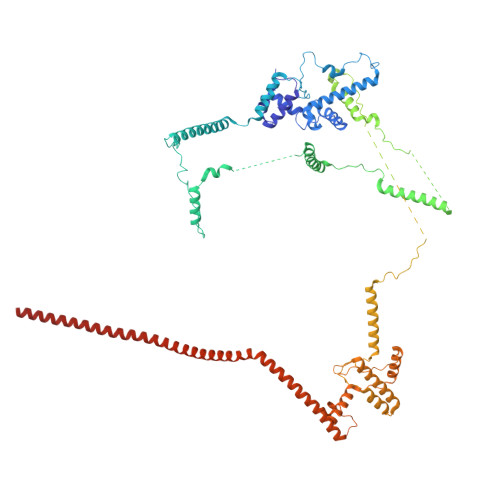
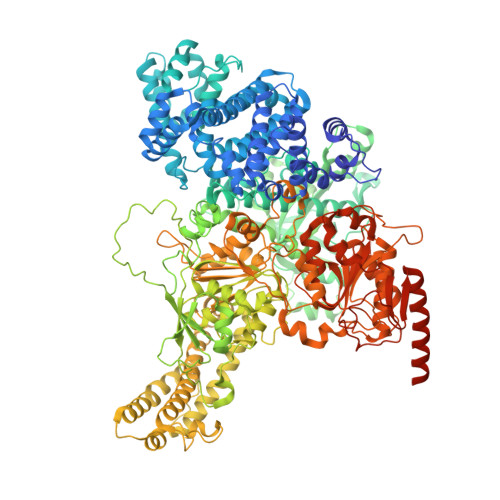
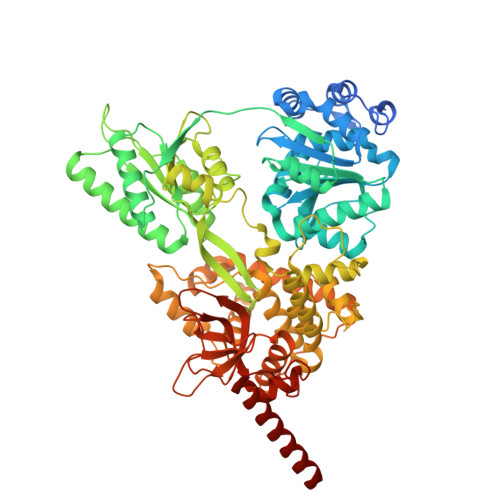
The spliceosome, a highly dynamic macromolecular assembly, catalyzes the precise removal of introns from pre-mRNAs. Recent studies have provided comprehensive structural insights into the step-wise assembly, catalytic splicing and final disassembly of the spliceosome. However, the molecular details of how the spliceosome recognizes and rejects suboptimal splicing substrates remained unclear. Here, we show cryo-electron microscopy structures of spliceosomal quality control complexes from a thermophilic eukaryote, Chaetomium thermophilum. The spliceosomes, henceforth termed B* Q , are stalled at a catalytically activated state but prior to the first splicing reaction due to an aberrant 5' splice site conformation. This state is recognized by G-patch protein GPATCH1, which is docked onto PRP8-EN and -RH domains and has recruited the cognate DHX35 helicase to its U2 snRNA substrate. In B* Q , DHX35 has dissociated the U2/branch site helix, while the disassembly helicase DHX15 is docked close to its U6 RNA 3'-end substrate. Our work thus provides mechanistic insights into the concerted action of two spliceosomal helicases in maintaining splicing fidelity by priming spliceosomes that are bound to aberrant splice substrates for disassembly.
- Minhang Hospital & Institutes of Biomedical Sciences, Shanghai Key Laboratory of Medical Epigenetics, International Co-laboratory of Medical Epigenetics and Metabolism, Fudan University, Shanghai, China.
Organizational Affiliation: