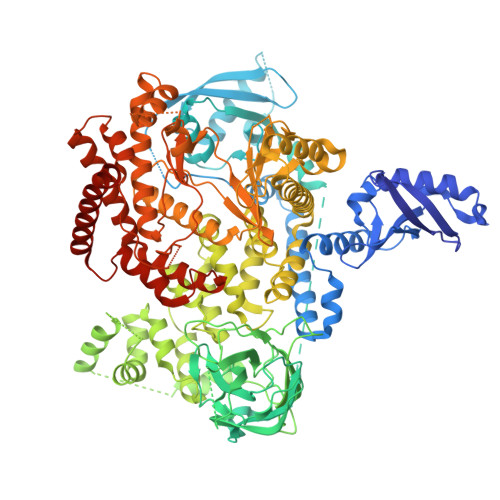
A long-lasting PI3K delta inhibitor zandelisib forms a water-shielded hydrogen bond with p110 delta and demonstrates sustained inhibitory effects.
Kunieda, K., Nagiri, C., Watanabe, M., Yoshida, T., Zou, J., Kaneda, A., Takahashi, Y., Aburai, K., Saito, J.I., Umehara, H., Otsu, Y., Ishii, T.(2025) Am J Cancer Res 15: 2097-2110
- PubMed: 40520883
- DOI: https://doi.org/10.62347/JCGQ1004
- Primary Citation of Related Structures:
9L3R - PubMed Abstract:
Phosphatidylinositol 3-kinase isoform δ (PI3Kδ) phosphorylates phosphatidylinositol lipids, activating the AKT signaling pathway, which is crucial for essential cellular functions in B cells. Zandelisib, a selective PI3Kδ inhibitor, is under clinical development for treating B cell malignancies. Its intermittent dosing regimen sustains therapeutic effects while minimizing adverse effects. We explored zandelisib's pharmacological activity, focusing on its long-lasting property as a PI3Kδ inhibitor. To gain mechanistic insights, we compared the crystal structure of PI3Kδ in complex with zandelisib with other PI3K inhibitors. The binding kinetics of zandelisib, parsaclisib, idelalisib, and duvelisib to PI3Kδ were evaluated using surface plasmon resonance (SPR) analysis with the Biacore TM system, and their binding in living cells was confirmed using the NanoBRET TM TE Intracellular Kinase Assay system. We assessed the effects of drug wash-out on intracellular drug concentrations, AKT phosphorylation inhibitory activity, and cell growth inhibitory activity in SU-DHL-6 or WSU-FSCCL B cell lymphoma cell lines. Pharmacokinetics/pharmacodynamics analysis and anti-tumor activity evaluation were performed in mice bearing SU-DHL-6 tumors. The binding mode of zandelisib to PI3Kδ was revealed by X-ray crystallography. SPR analysis showed that zandelisib had a slower dissociation rate than other compounds, which was confirmed in cell-based binding assays. Idelalisib, parsaclisib, and duvelisib lost their PI3Kδ inhibitory activity by wash-out and showed a decreased cell growth inhibitory activity. In comparison, zandelisib exhibited sustained inhibitory activity against PI3Kδ and showed a more gradual decrease in cell growth inhibitory activity. Drug concentration after wash-out was highest for zandelisib. In vivo experiments using SU-DHL-6 tumor-bearing mice found zandelisib sustained PI3Kδ inhibitory effects for 8 hours at 50 mg/kg and 24 hours at 100 mg/kg. These results reflected significant anti-tumor activity of zandelisib in the B cell lymphoma model. The crystal structure of PI3Kδ in complex with zandelisib was determined at 2.5 Å resolution, revealing the benzimidazole group in zandelisib formed a hydrogen bond to the side chain of Lys779 in p110δ, the catalytic subunit of PI3Kδ. These studies demonstrated a longer duration of action of zandelisib compared to the other compounds, which was attributable to the hydrogen bond between zandelisib and Lys779 in p110δ.
- Research Division, Kyowa Kirin Co., Ltd. 1188, Shimotogari, Nagaizumi-cho, Sunto-gun, Shizuoka, Japan.
Organizational Affiliation: