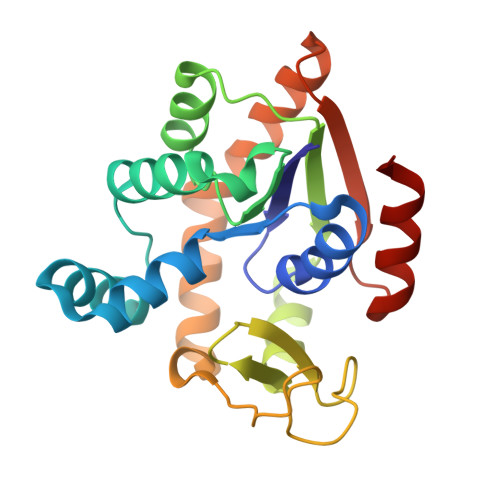
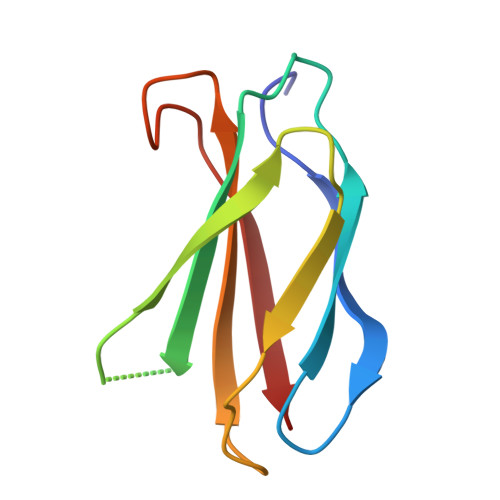
Binding mechanism of adenylate kinase-specific monobodies.
Nakamura, I., Amesaka, H., Nagao, S., Orito, N., Negi, S., Tanaka, S.I., Matsuo, T.(2025) FEBS Lett 599: 1948-1963
- PubMed: 40400134
- DOI: https://doi.org/10.1002/1873-3468.70076
- Primary Citation of Related Structures:
9L14 - PubMed Abstract:
Monobodies are synthetic antibody-mimetic proteins that regulate enzyme functions through protein-protein interactions. In this study, we investigated the binding mechanisms of monobodies to adenylate kinase (Adk). Calorimetric and X-ray crystallographic analyses revealed that CL-1, a monobody specific for the CLOSED form of Adk, binds to the CORE domain of Adk in an enthalpy-driven manner, forming several hydrogen bonds and a cation-π interaction at the protein interface, without perturbing the Adk backbone. In contrast, OP-4, an OPEN-form-specific monobody, exhibited entropy-driven binding. 1 H- 15 N 2D nuclear magnetic resonance (NMR), 31 P-NMR, and calorimetric studies revealed conformational perturbations to Adk by OP-4, while substrate access remained intact. The different thermodynamic and structural effects between the monobodies highlight the diverse binding mechanisms among monobodies.
- Division of Materials Science, Nara Institute of Science and Technology (NAIST), Ikoma, Nara, Japan.
Organizational Affiliation: