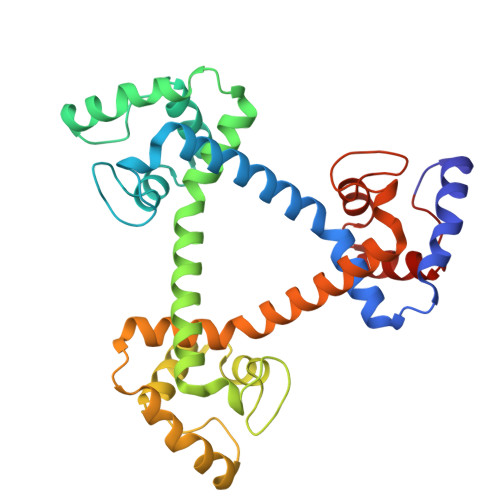
Construction of a Cyclic Regular-Triangle Trimer of Cytochrome c 555 with a Central Hole Using Sortase A.
Novientri, G., Fujiwara, K., Mashima, T., Matsuura, H., Ogata, H., Uchihashi, T., Fujii, S., Sambongi, Y., Hirota, S.(2025) Chemistry 31: e202404736-e202404736
- PubMed: 40148242
- DOI: https://doi.org/10.1002/chem.202404736
- Primary Citation of Related Structures:
9L08 - PubMed Abstract:
Protein-based supramolecules require precise arrangement of building blocks. A regular-triangle trimer (cp-c 555 ) 3 has been constructed using an α-helix-inserted-circular permutant (cp-c 555 ) of Aquifex aeolicus cytochrome (cyt) c 555 , where the trimers may dissociate to monomers. In this study, we stabilized the regular-triangle structure by constructing a cyclic regular-triangle of three α-helix-linked cyt c 555 molecules using sortase-mediated ligation (SML). Comparing SML using sortase A for six cp-c 555 variant trimers, the variant with GGG at the N-terminus and LPETG at the C-terminus reacted most efficiently. OP-(c 555 ) 3 was designed, in which two cyt c 555 molecules were fused using an α-helix, generating a dimer. The cyt c 555 C-terminal region was attached to the N-terminus of the dimer, and the cyt c 555 N-terminal region was attached to the C-terminus of the dimer using the same α-helix. OP-(c 555 ) 3 was expressed in Escherichia coli cells, and the termini were connected by SML, forming a cyclic regular-triangle, CL-(c 555 ) 3 . CL-(c 555 ) 3 showed higher thermostability than (cp-c 555 ) 3 and OP-(c 555 ) 3 . CL-(c 555 ) 3 structural stability was confirmed using high-speed atomic force microscopy. The X-ray crystal structure of CL-(c 555 ) 3 showed a cyclic structure and a nanoporous supramolecular assembly. These results demonstrate that a nanoporous supramolecular assembly can be constructed by designing a cyclic molecule with a central hole using SML.
- Division of Materials Science, Graduate School of Science and Technology, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara, Japan.
Organizational Affiliation: