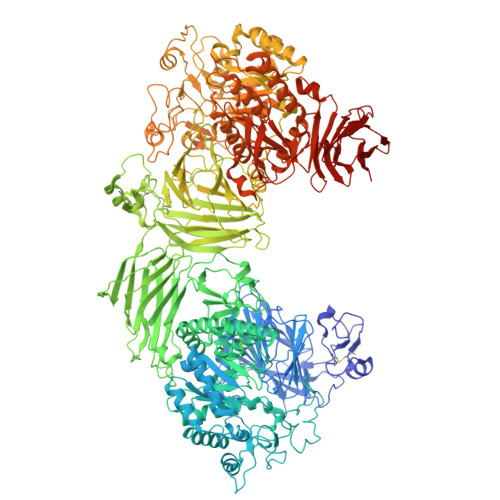
Porcine serum maltase-glucoamylase: structure, kinetics, and inhibition.
Watanabe, K., Tagami, T., Biwa, C., Kawasaki, M., Adachi, N., Moriya, T., Senda, T., Okuyama, M.(2026) J Enzyme Inhib Med Chem 41: 2612391-2612391
- PubMed: 41534875
- DOI: https://doi.org/10.1080/14756366.2025.2612391
- Primary Citation of Related Structures:
9KZ6, 9KZ7 - PubMed Abstract:
Maltase-glucoamylase (MGAM) is a small-intestinal enzyme comprising two tandem α-glucosidase units, NtMGAM and CtMGAM, each capable of hydrolysing maltodextrins into glucose. MGAM serves as a therapeutic target for managing postprandial hyperglycaemia; comprehensive insights into its full-length three-dimensional structure and inhibitor kinetics remains limited. Here, we demonstrate that the α-glucosidase in porcine serum is comparable to that encoded by the MGAM gene. Using cryo-electron microscopy, we determined the complex structure of serum MGAM with the inhibitor acarviosyl-maltotriose (AC5), which was found to bind exclusively to the active sites of each unit, confirming the presence of independent catalytic sites. AC5 was shown to exhibit mixed-type inhibition towards full-length serum MGAM and competitive inhibition against both recombinant NtMGAM and CtMGAM. The apparent mixed-type inhibition can be more accurately attributed to dual competitive inhibition mechanisms. These findings contribute to the advancement of functional foods and therapeutic interventions for postprandial hyperglycaemia and type 2 diabetes.
- Research Faculty of Agriculture, Hokkaido University, Sapporo, Hokkaido, Japan.
Organizational Affiliation: